The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: a bioinformatic comparison of Toc and Tic components in plants, green algae and red algae - PubMed (original) (raw)
Comparative Study
The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: a bioinformatic comparison of Toc and Tic components in plants, green algae and red algae
Ming Kalanon et al. Genetics. 2008 May.
Abstract
The recently completed genome of Chlamydomonas reinhardtii was surveyed for components of the chloroplast protein translocation complexes. Putative components were identified using reciprocal BlastP searches with the protein sequences of Arabidopsis thaliana as queries. As a comparison, we also surveyed the new genomes of the bryophyte Physcomitrella patens, two prasinophyte green algae (Ostreococcus lucimarinus and Ostreococcus tauri), the red alga Cyanidioschizon merolae, and several cyanobacteria. Overall, we found that the components of the import pathway are remarkably well conserved, particularly among the Viridiplantae lineages. Specifically, C. reinhardtii contained almost all the components found in A. thaliana, with two exceptions. Missing from C. reinhardtii are the C-terminal ferredoxin-NADPH-reductase (FNR) binding domain of Tic62 and a full-length, TPR-bearing Toc64. Further, the N-terminal domain of C. reinhardtii Toc34 is highly acidic, whereas the analogous region in C. reinhardtii Toc159 is not. This reversal of the vascular plant model may explain the similarity of C. reinhardtii chloroplast transit peptides to mitochondrial-targeting peptides. Other findings from our genome survey include the absence of Tic22 in both Ostreococcus genomes; the presence of only one Toc75 homolog in C. merolae; and, finally, a distinctive propensity for gene duplication in P. patens.
Figures
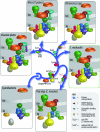
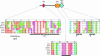
Figure 1.—
Evolution of the chloroplast protein translocation machinery. Components of Toc and Tic translocons of different phylogenetic lineages are shown, highlighting the overall continuity of the protein import machinery within red algae, chlorophytes, prasinophytes, bryophytes, and vascular plants. First, components derived from the cyanobacterial endosymbiont are also shown, including Omp85, Tic20, Tic22, Tic55, Tic32, and the NAD-binding domain of Tic62, as well as stromal factors such as ClpC. Components acquired early during plastid acquisition are represented in both the red and green lineages, including the conversion of Omp85 to Toc75, Tic110, Toc159, and Toc34. Tic40 was developed after the Viridiplantae diverged, since it is absent from red algae and cyanobacteria. Toc64 is also specific to Viridiplantae, but it is also absent from C. reinhardtii. It may have developed after the divergence of the chlorophytes or perhaps was lost specifically from the chlorophyte lineage. Specific to the vascular plant genomes is full-length Tic62, encoding a C-terminal FNR-binding domain. However, the N-terminal NAD-binding domain is found in all surveyed genomes, including cyanobacteria. Finally, Toc12 has been identified only in P. sativum to date.
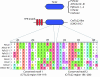
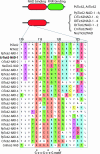
Figure 2.—
CrToc34 has a negatively charged N terminus. Toc34 alignment, indicating the dimerization motif (D1), arginine finger (R), and the predicted transmembrane helix (TM). The model shows the GTPase (green) and transmembrane helix (red) and highlights the length of and negative charges of the N terminus of CrToc34 (represented by “-”).
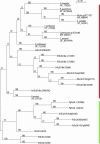
Figure 3.—
Phylogenetic tree of Tic20. A maximum-likelihood tree drawn from 125 characters with bootstrap values for 100 replicate trees shows that Tic20 proteins form two distinct clades. The first clade (green line) contains PsTic20 orthologs. Each Viridiplantae genome surveyed is represented here. The second clade (red line) contains the cyanobacterial homologs, as well as a subclade that contains Tic20-like proteins.
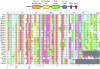
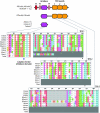
Figure 4.—
Conserved Tic22 motifs. Alignment of Tic22 proteins highlighting two conserved motifs. The Tic22 domain is represented in blue, while the TPR domains unique to CmTic22-like are shown in red.
Figure 5.—
Tic55 protein structure. Tic55 overview including the Rieske motif (orange), the mononuclear iron-binding site (yellow), the C-x-x-C motif (blue), two transmembrane domains (red), and the PFAM PAO motif (green). The alignment highlights conserved residues within these domains (black dots). Also indicated are positions where Tic55 orthologs encode a basic residue, instead of a proline at position +7 (red star), and an indel that is absent from Tic55 at position +16 (blue star).
Figure 6.—
Tic62 is an extended short-chain dehydrogenase. The conserved domains of full-length Tic62 are modeled with an N-terminal NAD-binding domain, which is classified within the “extended” class of short-chain dehydrogenases (red), and a C-terminal FNR-binding domain (green). Homologs from nonvascular genomes encode only the NAD-binding domain. The signature motif of extended SDR proteins is shown (G-x-x-G-x-x-G, black dots).
Figure 7.—
Tic32 is a classical short-chain dehydrogenase. Tic32 domains show the NAD-binding domain (red) and C-terminal 1-12 CaM-binding motif (purple). Indicated are the signature motif of classical SDR proteins (G-x-G-x-x-x-G) and catalytic tetrad residues (black dots). The majority of Viridiplantae sequences encode these residues in a Y-G-Q-S-K motif. A red box indicates the 1-12 CaM-binding motif, defined as bulky hydrophobic residues at positions +1 and +12.
Figure 8.—
Tic40 conserved motifs. The transmembrane domain (red), TPR motif (orange), and Hip/Hop domain (blue) of Tic40 proteins are shown. The putative processing site (W-h-G-h-G-h) is highlighted in a red box within the transmembrane domain. The conserved Y-P-Y-L-P-E motif within the TPR motif is also shown (black underlines), along with two chaperone-binding sites. Blue stars indicate the conserved asparagine/aspartic acid residues.
Figure 9.—
Toc64 conserved motifs. Domains of Toc64 are shown, including the amidase domain (purple), three TPR domains (orange), and transmembrane helices (red). Putative transmembrane domains are indicated by red outlining (Q
badou
et al. 2007). Highlighted is the catalytic site of the amidase (blue star), inactive glycine residues are shown in a red box and presumably active serine residues below. Alignments show that the TPR motifs are conserved, but are completely absent from CrToc64.
Similar articles
- Tic62: a protein family from metabolism to protein translocation.
Balsera M, Stengel A, Soll J, Bölter B. Balsera M, et al. BMC Evol Biol. 2007 Mar 20;7:43. doi: 10.1186/1471-2148-7-43. BMC Evol Biol. 2007. PMID: 17374152 Free PMC article. - Coexpressed subunits of dual genetic origin define a conserved supercomplex mediating essential protein import into chloroplasts.
Ramundo S, Asakura Y, Salomé PA, Strenkert D, Boone M, Mackinder LCM, Takafuji K, Dinc E, Rahire M, Crèvecoeur M, Magneschi L, Schaad O, Hippler M, Jonikas MC, Merchant S, Nakai M, Rochaix JD, Walter P. Ramundo S, et al. Proc Natl Acad Sci U S A. 2020 Dec 22;117(51):32739-32749. doi: 10.1073/pnas.2014294117. Epub 2020 Dec 3. Proc Natl Acad Sci U S A. 2020. PMID: 33273113 Free PMC article. - Reconstructing the mitochondrial protein import machinery of Chlamydomonas reinhardtii.
Figueroa-Martínez F, Funes S, Franzén LG, González-Halphen D. Figueroa-Martínez F, et al. Genetics. 2008 May;179(1):149-55. doi: 10.1534/genetics.108.087965. Genetics. 2008. PMID: 18493047 Free PMC article. - The chloroplast protein import system: from algae to trees.
Shi LX, Theg SM. Shi LX, et al. Biochim Biophys Acta. 2013 Feb;1833(2):314-31. doi: 10.1016/j.bbamcr.2012.10.002. Epub 2012 Oct 9. Biochim Biophys Acta. 2013. PMID: 23063942 Review. - Genome analysis and its significance in four unicellular algae, Cyanidioschyzon [corrected] merolae, Ostreococcus tauri, Chlamydomonas reinhardtii, and Thalassiosira pseudonana.
Misumi O, Yoshida Y, Nishida K, Fujiwara T, Sakajiri T, Hirooka S, Nishimura Y, Kuroiwa T. Misumi O, et al. J Plant Res. 2008 Jan;121(1):3-17. doi: 10.1007/s10265-007-0133-9. Epub 2007 Dec 12. J Plant Res. 2008. PMID: 18074102 Review.
Cited by
- Folding and self-association of atTic20 in lipid membranes: implications for understanding protein transport across the inner envelope membrane of chloroplasts.
Campbell JH, Hoang T, Jelokhani-Niaraki M, Smith MD. Campbell JH, et al. BMC Biochem. 2014 Dec 31;15:29. doi: 10.1186/s12858-014-0029-y. BMC Biochem. 2014. PMID: 25551276 Free PMC article. - The physical and functional borders of transit peptide-like sequences in secondary endosymbionts.
Felsner G, Sommer MS, Maier UG. Felsner G, et al. BMC Plant Biol. 2010 Oct 19;10:223. doi: 10.1186/1471-2229-10-223. BMC Plant Biol. 2010. PMID: 20958984 Free PMC article. - Evidence for the retention of two evolutionary distinct plastids in dinoflagellates with diatom endosymbionts.
Hehenberger E, Imanian B, Burki F, Keeling PJ. Hehenberger E, et al. Genome Biol Evol. 2014 Sep;6(9):2321-34. doi: 10.1093/gbe/evu182. Genome Biol Evol. 2014. PMID: 25172904 Free PMC article. - An Rh1-GFP fusion protein is in the cytoplasmic membrane of a white mutant strain of Chlamydomonas reinhardtii.
Yoshihara C, Inoue K, Schichnes D, Ruzin S, Inwood W, Kustu S. Yoshihara C, et al. Mol Plant. 2008 Nov;1(6):1007-20. doi: 10.1093/mp/ssn074. Epub 2008 Nov 14. Mol Plant. 2008. PMID: 19825599 Free PMC article. - Treasure hunting in the Chlamydomonas genome.
Vallon O, Dutcher S. Vallon O, et al. Genetics. 2008 May;179(1):3-6. doi: 10.1534/genetics.104.179101. Genetics. 2008. PMID: 18493035 Free PMC article. No abstract available.
References
- Aronsson, H., P. Boij, R. Patel, A. Wardle, M. Töpel et al., 2007. Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 52 53–68. - PubMed
- Baldwin, A. J., and K. Inoue, 2006. The most C-terminal tri-glycine segment within the polyglycine stretch of the pea Toc75 transit peptide plays a critical role for targeting the protein to the chloroplast outer envelope membrane. FEBS J. 273 1547–1555. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials