Alterations in mossy fiber physiology and GAP-43 expression and function in transgenic mice overexpressing HuD - PubMed (original) (raw)
Alterations in mossy fiber physiology and GAP-43 expression and function in transgenic mice overexpressing HuD
Daniel C Tanner et al. Hippocampus. 2008.
Abstract
HuD is a neuronal RNA-binding protein associated with the stabilization of mRNAs for GAP-43 and other neuronal proteins that are important for nervous system development and learning and memory mechanisms. To better understand the function of this protein, we generated transgenic mice expressing human HuD (HuD-Tg) in adult forebrain neurons. We have previously shown that expression of HuD in adult dentate granule cells results in an abnormal accumulation of GAP-43 mRNA via posttranscriptional mechanisms. Here we show that this mRNA accumulation leads to the ectopic expression of GAP-43 protein in mossy fibers. Electrophysiological analyses of the mossy fiber to CA3 synapse of HuD-Tg mice revealed increases in paired-pulse facilitation (PPF) at short interpulse intervals and no change in long-term potentiation (LTP). Presynaptic calcium transients at the same synapses exhibited faster time constants of decay, suggesting a decrease in the endogenous Ca(2+) buffer capacity of mossy fiber terminals of HuD-Tg mice. Under resting conditions, GAP-43 binds very tightly to calmodulin sequestering it and then releasing it upon PKC-dependent phosphorylation. Therefore, subsequent studies examined the extent of GAP-43 phosphorylation and its association to calmodulin. We found that despite the increased GAP-43 expression in HuD-Tg mice, the levels of PKC-phosphorylated GAP-43 were decreased in these animals. Furthermore, in agreement with the increased proportion of nonphosphorylated GAP-43, HuD-Tg mice showed increased binding of calmodulin to this protein. These results suggest that a significant amount of calmodulin may be trapped in an inactive state, unable to bind free calcium, and activate downstream signaling pathways. In conclusion, we propose that an unregulated expression of HuD disrupts mossy fiber physiology in adult mice in part by altering the expression and phosphorylation of GAP-43 and the amount of free calmodulin available at the synaptic terminal.
(c) 2008 Wiley-Liss, Inc.
Figures
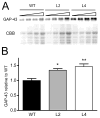
Figure 1. Increased levels of GAP-43 in the hippocampus of HuD-Tg mice
The levels of GAP-43 were determined in hippocampal homogenates from adult HuD-Tg mice of line 2 and line 4 and from non-transgenic littermates (WT) by western blots. (A) Panels show GAP-43 protein levels and CBB staining of serial dilutions of hippocampal extracts. (B) The optical densities of GAP-43 were corrected by that of CBB. Both lines of HuD-Tg mice show significant increases in GAP-43 expression (N=9 *p<0.05 and **p<0.01, one-way ANOVA).
Figure 2. GAP-43 protein is present in mossy fiber terminals of HuD transgenic mice but not in wild type mice
GAP-43 immunofluorescence (green, A, D,G) Calbindin D28K ( red, B,E,H) and merged images (C,F,I) show the increased expression of GAP-43 and its colocalization with calbindin in HuD-Tg mice. Images shown in the top panels were taken using a 4X objective and those in the bottom panels were taken at 20X. Note that although wild type mice only show background GAP-43 staining in mossy fibers (A and J), both lines of HuD-Tg mice (M, P) express high levels of this protein (D, F, K, P). Slm, stratum lacunosum moleculare; sml, stratum moleculare; sl, stratum lucidum; so, stratum oriens; sr, stratum radiatum, and sp, stratum pyramidale. Scale bars = 300 μm (A–I) and 50 μm (J–R).
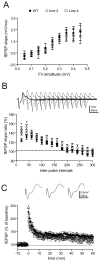
Figure 3. Enhanced paired pulse facilitation in mossy fiber terminals of HuD-Tg mice
A) The input-output relationships of mossy fibers, as represented by the presynaptic fiber volley amplitude (binned in 0.05 mV) vs. fEPSP slopes, did not reveal significant changes in either genotypes (two-way ANOVA, p>0.05 for genotype effects). B) Altered HuD and GAP-43 expression leads to increased paired pulse facilitation in mossy fiber-CA3 synapses at short interpulse intervals (two-way ANOVA, with Bonferroni post hoc tests. *p<0.05). C) No changes of tetanus-induced, NMDAR-independent LTP in either transgenic line. Mossy fiber LTP was induced by four 1-sec trains of 100Hz stimuli that were separated by a 20-sec inter pulse interval. 50 uM AP5 was present during the experimental duration. No significant changes were obtained at 60 min post-tetanus (one-way ANOVA p>0.05). All the electrophysiological measurements used 7–8 mice per group. Filled circles, WT; open circles, line 2; open squares, Line 4
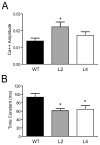
Figure 4. Changes in residual calcium levels and dynamics in mossy fibers of HuD Tg-mice
Calcium transients were measured in distal mossy fibers after proximal stimulation using Mg Green as described in the Materials and Methods. Animals of line 2 showed increased in the initial amplitude of the calcium signal (A) whereas both lines of HuD-Tg mice exhibited decreases in the time constant of decay (B). *p<0.05.
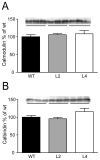
Figure 5. No differences in the total levels of the calcium-binding proteins calmodulin and calbindin in the hippocampus of HuD-Tg mice
Protein levels were determined by western blots in the S1 fraction as described in the Methods. Results are representative from three independent experiments using 5 animals per group.
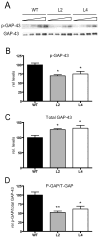
Figure 6. Decreased levels of PKC-phosphorylated GAP-43 in HuD-Tg mice
The levels of phosphorylated GAP-43 in hippocampal homogenates were determined using a phospho-specific antibody. A) Western blots showing the levels of phosphorylated and total GAP-43 protein increased in HuD-Tg mice. (B–C). The levels of phosphorylated GAP-43 (p-GAP) and total GAP-43 (T-GAP) were corrected by Coomassie blue staining. (D) The ratios of p-GAP-43 to total GAP-43 were calculated using the intensities of the bands corresponding to these proteins on the same blots. *p<0.05**p<0.01, N=5 animals per group.
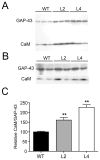
Figure 7. Increased binding of calmodulin to GAP-43 in HuD-Tg mice
Western blots show the levels of calmodulin and GAP-43 in the membrane fraction (A) and after immunoprecipitation with GAP-43 specific antibodies (B). The amount of calmodulin bound to GAP-43 increased significantly in both lines of HuD-Tg mice (C). Results are representative of three independent experiments run in duplicates using 3 animals from each group. **p<0.01
Similar articles
- In vivo post-transcriptional regulation of GAP-43 mRNA by overexpression of the RNA-binding protein HuD.
Bolognani F, Tanner DC, Merhege M, Deschênes-Furry J, Jasmin B, Perrone-Bizzozero NI. Bolognani F, et al. J Neurochem. 2006 Feb;96(3):790-801. doi: 10.1111/j.1471-4159.2005.03607.x. Epub 2006 Jan 9. J Neurochem. 2006. PMID: 16405504 - Increased expression of axogenesis-related genes and mossy fibre length in dentate granule cells from adult HuD overexpressor mice.
Perrone-Bizzozero NI, Tanner DC, Mounce J, Bolognani F. Perrone-Bizzozero NI, et al. ASN Neuro. 2011;3(5):259-70. doi: 10.1042/AN20110015. ASN Neuro. 2011. PMID: 22004431 Free PMC article. - Coordinated expression of HuD and GAP-43 in hippocampal dentate granule cells during developmental and adult plasticity.
Bolognani F, Tanner DC, Nixon S, Okano HJ, Okano H, Perrone-Bizzozero NI. Bolognani F, et al. Neurochem Res. 2007 Dec;32(12):2142-51. doi: 10.1007/s11064-007-9388-8. Epub 2007 Jun 19. Neurochem Res. 2007. PMID: 17577668 - Emerging complexity of the HuD/ELAVl4 gene; implications for neuronal development, function, and dysfunction.
Bronicki LM, Jasmin BJ. Bronicki LM, et al. RNA. 2013 Aug;19(8):1019-37. doi: 10.1261/rna.039164.113. RNA. 2013. PMID: 23861535 Free PMC article. Review. - Role of HuD in nervous system function and pathology.
Perrone-Bizzozero N, Bird CW. Perrone-Bizzozero N, et al. Front Biosci (Schol Ed). 2013 Jan 1;5(2):554-63. doi: 10.2741/s389. Front Biosci (Schol Ed). 2013. PMID: 23277068 Review.
Cited by
- miR-375 inhibits differentiation of neurites by lowering HuD levels.
Abdelmohsen K, Hutchison ER, Lee EK, Kuwano Y, Kim MM, Masuda K, Srikantan S, Subaran SS, Marasa BS, Mattson MP, Gorospe M. Abdelmohsen K, et al. Mol Cell Biol. 2010 Sep;30(17):4197-210. doi: 10.1128/MCB.00316-10. Epub 2010 Jun 28. Mol Cell Biol. 2010. PMID: 20584986 Free PMC article. - HuD Binds to and Regulates Circular RNAs Derived From Neuronal Development- and Synaptic Plasticity-Associated Genes.
Dell'Orco M, Oliver RJ, Perrone-Bizzozero N. Dell'Orco M, et al. Front Genet. 2020 Aug 5;11:790. doi: 10.3389/fgene.2020.00790. eCollection 2020. Front Genet. 2020. PMID: 32849796 Free PMC article. - The RNA-Binding Protein HuD Regulates Alternative Splicing and Alternative Polyadenylation in the Mouse Neocortex.
Sena RM, Twiss JL, Gardiner AS, Dell'Orco M, Linsenbardt DN, Perrone-Bizzozero NI. Sena RM, et al. Molecules. 2021 May 11;26(10):2836. doi: 10.3390/molecules26102836. Molecules. 2021. PMID: 34064652 Free PMC article. - Neuronal RNA-binding protein HuD regulates addiction-related gene expression and behavior.
Oliver RJ, Brigman JL, Bolognani F, Allan AM, Neisewander JL, Perrone-Bizzozero NI. Oliver RJ, et al. Genes Brain Behav. 2018 Apr;17(4):e12454. doi: 10.1111/gbb.12454. Epub 2018 Jan 26. Genes Brain Behav. 2018. PMID: 29283498 Free PMC article. - A systems level, functional genomics analysis of chronic epilepsy.
Winden KD, Karsten SL, Bragin A, Kudo LC, Gehman L, Ruidera J, Geschwind DH, Engel J Jr. Winden KD, et al. PLoS One. 2011;6(6):e20763. doi: 10.1371/journal.pone.0020763. Epub 2011 Jun 14. PLoS One. 2011. PMID: 21695113 Free PMC article.
References
- Akers RF, Routtenberg A. Protein kinase C phosphorylates a 47 Mr protein (F1) directly related to synaptic plasticity. Brain Res. 1985;334(1):147–51. - PubMed
- Alexander KA, Cimler BM, Meier KE, Storm DR. Regulation of calmodulin binding to P-57. A neurospecific calmodulin binding protein. J Biol Chem. 1987;262(13):6108–13. - PubMed
- Anderson KD, Morin MA, Beckel-Mitchener A, Mobarak CD, Neve RL, Furneaux HM, Burry R, Perrone-Bizzozero NI. Overexpression of HuD, but not of its truncated form HuD I+II, promotes GAP-43 gene expression and neurite outgrowth in PC12 cells in the absence of nerve growth factor. J Neurochem. 2000;75(3):1103–14. - PubMed
- Anderson KD, Sengupta J, Morin M, Neve RL, Valenzuela CF, Perrone-Bizzozero NI. Overexpression of HuD accelerates neurite outgrowth and increases GAP-43 mRNA expression in cortical neurons and retinoic acid-induced embryonic stem cells in vitro. Exp Neurol. 2001;168(2):250–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG022574-05/AG/NIA NIH HHS/United States
- R01 NS030255/NS/NINDS NIH HHS/United States
- T32 AA1427/AA/NIAAA NIH HHS/United States
- NS30255/NS/NINDS NIH HHS/United States
- R01 AG022574/AG/NIA NIH HHS/United States
- AG022574/AG/NIA NIH HHS/United States
- R01 NS030255-16/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous