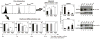Transcriptome-wide noise controls lineage choice in mammalian progenitor cells - PubMed (original) (raw)
Transcriptome-wide noise controls lineage choice in mammalian progenitor cells
Hannah H Chang et al. Nature. 2008.
Abstract
Phenotypic cell-to-cell variability within clonal populations may be a manifestation of 'gene expression noise', or it may reflect stable phenotypic variants. Such 'non-genetic cell individuality' can arise from the slow fluctuations of protein levels in mammalian cells. These fluctuations produce persistent cell individuality, thereby rendering a clonal population heterogeneous. However, it remains unknown whether this heterogeneity may account for the stochasticity of cell fate decisions in stem cells. Here we show that in clonal populations of mouse haematopoietic progenitor cells, spontaneous 'outlier' cells with either extremely high or low expression levels of the stem cell marker Sca-1 (also known as Ly6a; ref. 9) reconstitute the parental distribution of Sca-1 but do so only after more than one week. This slow relaxation is described by a gaussian mixture model that incorporates noise-driven transitions between discrete subpopulations, suggesting hidden multi-stability within one cell type. Despite clonality, the Sca-1 outliers had distinct transcriptomes. Although their unique gene expression profiles eventually reverted to that of the median cells, revealing an attractor state, they lasted long enough to confer a greatly different proclivity for choosing either the erythroid or the myeloid lineage. Preference in lineage choice was associated with increased expression of lineage-specific transcription factors, such as a >200-fold increase in Gata1 (ref. 10) among the erythroid-prone cells, or a >15-fold increased PU.1 (Sfpi1) (ref. 11) expression among myeloid-prone cells. Thus, clonal heterogeneity of gene expression level is not due to independent noise in the expression of individual genes, but reflects metastable states of a slowly fluctuating transcriptome that is distinct in individual cells and may govern the reversible, stochastic priming of multipotent progenitor cells in cell fate decision.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
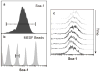
Figure 1. Robust clonal heterogeneity
a, b, Heterogeneity in Sca-1 expression among clonal cells (a) was significantly larger than the resolution limit of flow cytometry approximated by measurement of reference MESF beads (b). c, Stability of clonal heterogeneity in Sca-1 over 3 weeks.
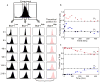
Figure 2. Restoration of heterogeneity from sorted cell fractions
a, Clonal cells with the highest (Sca-1High), middle (Sca-1Mid) and lowest (Sca-1Low) 15% Sca-1 expression independently re-established the parental extent of clonal heterogeneity after 216 h in separate culture. As an example, each cell in the Sca-1High experiment was theoretically partitioned into one of two GMM-subpopulations (blue and red). b, c, The temporal evolution of the means μ1,2 (b) and weights w1,2 (c) for the Sca-1High GMM subpopulations 1 and 2. The evolution of the weights was fitted to a sigmoidal function (c, dotted curves). Black dotted dash lines, equilibrium values for μı and wi.
Figure 3. Clonal heterogeneity governs differentiation potential
a–f, Sca-1Low (Low, black), Sca-1Mid (Mid, grey), and Sca-1High (High, white) fractions (a) stimulated by Epo (b) and GM-CSF (f) immediately after isolation showed variable differentiation rates into the erythroid and myeloid lineages, respectively. Upon 7, 14, and 21 days (d) of post-sort culture, Epo- treated cells showed convergence in both pre-stimulation, baseline Sca-1 expression (Fig. 2a) and relative differentiation rates (b–e). Asterisk, p < 0.001 (two-tailed normal-theory test). g, h, qRT-PCR analysis of GATA1 (g) and PU.1 (h) mRNA levels in Sca-1 sorted fractions. Means ± s.e.m. of triplicates shown; triple asterisk p < 10−5, double asterisk p < 0.0002, asterisk p < 0.003 (one-tail Student’s t-test). i, j, Western blot analysis of GATA1 (i) and PU.1 (j) protein levels in Sca-1 fractions (lanes 3–5) and mock-sorted cells (lane 6). MEL cell line (lane 1), positive control; G1E and 503 (lane 2) cell lines, negative controls for GATA1 and PU.1, respectively. GAPDH, loading control.
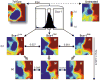
Figure 4. Clonal heterogeneity of Sca-1 expression reflects transcriptome-wide noise
Self-organizing maps of global gene expression for a subset of 2997 genes visualized with the GEDI program for Sca-1Low (L), Sca-1Mid (M), Sca-1High (H) fractions at 0 and 6 days (d) after FACS isolation and for a differentiated erythroid culture (7d Epo) and an untreated (Untreated) control sample. Pixels in the same location within each GEDI map contain the same minicluster of genes. Color of pixels indicates centroid value of gene expression level for each minicluster in log10 units of signal. Dissimilarity between transcriptomes indicated above  . GATA1-containing pixel boxed in white.
. GATA1-containing pixel boxed in white.
Similar articles
- Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios.
Hoppe PS, Schwarzfischer M, Loeffler D, Kokkaliaris KD, Hilsenbeck O, Moritz N, Endele M, Filipczyk A, Gambardella A, Ahmed N, Etzrodt M, Coutu DL, Rieger MA, Marr C, Strasser MK, Schauberger B, Burtscher I, Ermakova O, Bürger A, Lickert H, Nerlov C, Theis FJ, Schroeder T. Hoppe PS, et al. Nature. 2016 Jul 14;535(7611):299-302. doi: 10.1038/nature18320. Nature. 2016. PMID: 27411635 - Towards an understanding of lineage specification in hematopoietic stem cells: a mathematical model for the interaction of transcription factors GATA-1 and PU.1.
Roeder I, Glauche I. Roeder I, et al. J Theor Biol. 2006 Aug 21;241(4):852-65. doi: 10.1016/j.jtbi.2006.01.021. Epub 2006 Feb 28. J Theor Biol. 2006. PMID: 16510158 - Lineage marker synchrony in hematopoietic genealogies refutes the PU.1/GATA1 toggle switch paradigm.
Strasser MK, Hoppe PS, Loeffler D, Kokkaliaris KD, Schroeder T, Theis FJ, Marr C. Strasser MK, et al. Nat Commun. 2018 Jul 12;9(1):2697. doi: 10.1038/s41467-018-05037-3. Nat Commun. 2018. PMID: 30002371 Free PMC article. - The importance of PU.1 concentration in hematopoietic lineage commitment and maturation.
Dahl R, Simon MC. Dahl R, et al. Blood Cells Mol Dis. 2003 Sep-Oct;31(2):229-33. doi: 10.1016/s1079-9796(03)00152-9. Blood Cells Mol Dis. 2003. PMID: 12972030 Review. - Myeloid lineage commitment from the hematopoietic stem cell.
Iwasaki H, Akashi K. Iwasaki H, et al. Immunity. 2007 Jun;26(6):726-40. doi: 10.1016/j.immuni.2007.06.004. Immunity. 2007. PMID: 17582345 Review.
Cited by
- MONITTR allows real-time imaging of transcription and endogenous proteins in C. elegans.
Liu X, Chang Z, Sun P, Cao B, Wang Y, Fang J, Pei Y, Chen B, Zou W. Liu X, et al. J Cell Biol. 2025 Jan 6;224(1):e202403198. doi: 10.1083/jcb.202403198. Epub 2024 Oct 14. J Cell Biol. 2025. PMID: 39400293 - Discovery of prognostic lncRNAs in colorectal cancer using spatial transcriptomics.
Pinkney HR, Ross CR, Hodgson TO, Pattison ST, Diermeier SD. Pinkney HR, et al. NPJ Precis Oncol. 2024 Oct 10;8(1):230. doi: 10.1038/s41698-024-00728-1. NPJ Precis Oncol. 2024. PMID: 39390212 Free PMC article. - Irreversibility in bacterial regulatory networks.
Zhao Y, Wytock TP, Reynolds KA, Motter AE. Zhao Y, et al. Sci Adv. 2024 Aug 30;10(35):eado3232. doi: 10.1126/sciadv.ado3232. Epub 2024 Aug 28. Sci Adv. 2024. PMID: 39196926 Free PMC article. - Modeling relaxation experiments with a mechanistic model of gene expression.
Estavoyer M, Dufeu M, Ranson G, Lefort S, Voeltzel T, Maguer-Satta V, Gandrillon O, Lepoutre T. Estavoyer M, et al. BMC Bioinformatics. 2024 Aug 20;25(1):270. doi: 10.1186/s12859-024-05816-4. BMC Bioinformatics. 2024. PMID: 39164646 Free PMC article. - Identification of molecular determinants of gene-specific bursting patterns by high-throughput imaging screens.
Sood V, Holewinski R, Andresson T, Larson DR, Misteli T. Sood V, et al. bioRxiv [Preprint]. 2024 Jun 8:2024.06.08.597999. doi: 10.1101/2024.06.08.597999. bioRxiv. 2024. PMID: 38903099 Free PMC article. Preprint.
References
- Blake WJ, et al. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. - PubMed
- Elowitz MB, et al. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. - PubMed
- Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965–1969. - PubMed
- Rosenfeld N, et al. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials