Evolutionary origin of regulatory regions of retrogenes in Drosophila - PubMed (original) (raw)
Evolutionary origin of regulatory regions of retrogenes in Drosophila
Yongsheng Bai et al. BMC Genomics. 2008.
Abstract
Background: Retrogenes are processed copies of other genes. This duplication mechanism produces a copy of the parental gene that should not contain introns, and usually does not contain cis-regulatory regions. Here, we computationally address the evolutionary origin of promoter and other cis-regulatory regions in retrogenes using a total of 94 Drosophila retroposition events we recently identified. Previous tissue expression data has revealed that a large fraction of these retrogenes are specifically and/or highly expressed in adult testes of Drosophila.
Results: In this work, we infer that retrogenes do not generally carry regulatory regions from aberrant upstream or normal transcripts of their parental genes, and that expression patterns of neighboring genes are not consistently shared by retrogenes. Additionally, transposable elements do not appear to substantially provide regulatory regions to retrogenes. Interestingly, we find that there is an excess of retrogenes in male testis neighborhoods that is not explained by insertional biases of the retroelement machinery used for retroposition.
Conclusion: We conclude that retrogenes' regulatory regions mostly do not represent a random set of existing regulatory regions. On the contrary, our conclusion is that selection is likely to have played an important role in the persistence of autosomal testis biased retrogenes. Selection in favor of retrogenes inserted in male testis neighborhoods and at the sequence level to produce testis expression is postulated to have occurred.
Figures
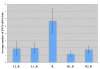
Figure 1
Adult testes expression level for the retrogene (R), and two flanking genes on each side (left (L) and right (R)). Standard error bars are given.
Similar articles
- Quality of regulatory elements in Drosophila retrogenes.
Bai Y, Casola C, Betrán E. Bai Y, et al. Genomics. 2009 Jan;93(1):83-9. doi: 10.1016/j.ygeno.2008.09.006. Epub 2008 Oct 23. Genomics. 2009. PMID: 18848618 Free PMC article. - Relocation facilitates the acquisition of short cis-regulatory regions that drive the expression of retrogenes during spermatogenesis in Drosophila.
Sorourian M, Kunte MM, Domingues S, Gallach M, Özdil F, Río J, Betrán E. Sorourian M, et al. Mol Biol Evol. 2014 Aug;31(8):2170-80. doi: 10.1093/molbev/msu168. Epub 2014 May 22. Mol Biol Evol. 2014. PMID: 24855141 Free PMC article. - Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila.
Bai Y, Casola C, Feschotte C, Betrán E. Bai Y, et al. Genome Biol. 2007;8(1):R11. doi: 10.1186/gb-2007-8-1-r11. Genome Biol. 2007. PMID: 17233920 Free PMC article. - Reverse transcriptase: mediator of genomic plasticity.
Brosius J, Tiedge H. Brosius J, et al. Virus Genes. 1995;11(2-3):163-79. doi: 10.1007/BF01728656. Virus Genes. 1995. PMID: 8828143 Review. - Modern genomes with retro-look: retrotransposed elements, retroposition and the origin of new genes.
Volff JN, Brosius J. Volff JN, et al. Genome Dyn. 2007;3:175-190. doi: 10.1159/000107611. Genome Dyn. 2007. PMID: 18753792 Review.
Cited by
- Noncoding sequences near duplicated genes evolve rapidly.
Kostka D, Hahn MW, Pollard KS. Kostka D, et al. Genome Biol Evol. 2010;2:518-33. doi: 10.1093/gbe/evq037. Epub 2010 Jun 29. Genome Biol Evol. 2010. PMID: 20660939 Free PMC article. - Nuclear transport genes recurrently duplicate by means of RNA intermediates in Drosophila but not in other insects.
Mirsalehi A, Markova DN, Eslamieh M, Betrán E. Mirsalehi A, et al. BMC Genomics. 2021 Dec 5;22(1):876. doi: 10.1186/s12864-021-08170-4. BMC Genomics. 2021. PMID: 34863092 Free PMC article. - Retrogenes in rice (Oryza sativa L. ssp. japonica) exhibit correlated expression with their source genes.
Sakai H, Mizuno H, Kawahara Y, Wakimoto H, Ikawa H, Kawahigashi H, Kanamori H, Matsumoto T, Itoh T, Gaut BS. Sakai H, et al. Genome Biol Evol. 2011;3:1357-68. doi: 10.1093/gbe/evr111. Epub 2011 Oct 31. Genome Biol Evol. 2011. PMID: 22042334 Free PMC article. - Evolution of three parent genes and their retrogene copies in Drosophila species.
O'Neill RS, Clark DV. O'Neill RS, et al. Int J Evol Biol. 2013;2013:693085. doi: 10.1155/2013/693085. Epub 2013 Jun 5. Int J Evol Biol. 2013. PMID: 23841016 Free PMC article. - Chimeric genes as a source of rapid evolution in Drosophila melanogaster.
Rogers RL, Hartl DL. Rogers RL, et al. Mol Biol Evol. 2012 Feb;29(2):517-29. doi: 10.1093/molbev/msr184. Epub 2011 Jul 18. Mol Biol Evol. 2012. PMID: 21771717 Free PMC article.
References
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, Ainscough R, Almeida JP, Babbage A, Bagguley C, Bailey J, Barlow K, Bates KN, Beasley O, Bird CP, Blakey S, Bridgeman AM, Buck D, Burgess J, Burrill WD, O'Brien KP, et al. The DNA sequence of human chromosome 22 [see comments] [published erratum appears in Nature 2000 Apr 20;404(6780):904] Nature. 1999;402:489–495. doi: 10.1038/990031. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases