Reversible cytoplasmic localization of the proteasome in quiescent yeast cells - PubMed (original) (raw)
Reversible cytoplasmic localization of the proteasome in quiescent yeast cells
Damien Laporte et al. J Cell Biol. 2008.
Abstract
The 26S proteasome is responsible for the controlled proteolysis of a vast number of proteins, including crucial cell cycle regulators. Accordingly, in Saccharomyces cerevisiae, 26S proteasome function is mandatory for cell cycle progression. In budding yeast, the 26S proteasome is assembled in the nucleus, where it is localized throughout the cell cycle. We report that upon cell entry into quiescence, proteasome subunits massively relocalize from the nucleus into motile cytoplasmic structures. We further demonstrate that these structures are proteasome cytoplasmic reservoirs that are rapidly mobilized upon exit from quiescence. Therefore, we have named these previously unknown structures proteasome storage granules (PSGs). Finally, we observe conserved formation and mobilization of these PSGs in the evolutionary distant yeast Schizosaccharomyces pombe. This conservation implies a broad significance for these proteasome reserves.
Figures
Figure 1.
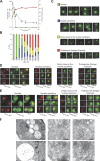
Proteasome relocalization upon entry into quiescence. (A–C) Cells expressing Pre6p-GFP were grown in YPDA medium at 30°C. (A) At various stages of growth, OD 600 nm was monitored (red curve), glucose concentration in the medium was measured (green curve), and the budding index was determined (blue curve; n > 200 cells for each time point). (B) Localization of Pre6p-GFP. For each time point, the localization of Pre6p-GFP fluorescence was scored as nuclear (green bar), at the nuclear periphery (blue bar), immobile dots close to the nuclear periphery (yellow bar), or bright mobile cytoplasmic dots (PSG) with or without nuclear periphery staining (red bar; n > 200 cells for each time point; two experiments; error bars show SD). The gray bars represent the percentage of cells in which GFP fluorescence was not detectable. (C) Typical images of each Pre6p-GFP localization patterns are shown. Images are maximal projection of 0.2-μm z stacks. Of note, for localization of Pre6p-GFP at the nuclear periphery, images were taken with an exposure time that was three times longer than for the other images. Furthermore, levels of gray of the top two rows of images range from 0 to 800 and the levels of gray of the two bottom rows of images range from 0 to 3,000. (D) Colocalization of the proteasome (as revealed by the 20S CP subunit Pup1p fused to RFP) and the nuclear envelope (as revealed by the nuclear pore complex protein Nup2p fused to GFP). (E) Colocalization of the proteasome (as revealed by the 20S CP subunit Pup1p fused to RFP) and the endoplasmic reticulum (as revealed by the membrane protein Pho88p fused to GFP). Images in D and E are single focal planes. Bars, 2 μm. (F) Immunolocalization using anti-20S CP antibodies detected with secondary antibodies linked to 10-nm gold particles in wild-type yeast cells grown for 4 d at 30°C in YPDA medium.
Figure 2.
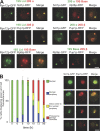
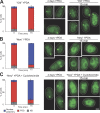
19S and 20S proteasome subunits colocalization. (A) Colocalization of 19S RP and 20S CP proteasome subunits in quiescent yeast cells. Cells coexpressing the indicated fusion proteins were imaged after 4 d of growth in YPDA at 30°C. GFP fluorescence is green and RFP fluorescence is red. Images are single focal planes. (B) Cells expressing Scl1p-GFP and Pup1p-RFP were grown in YPDA medium at 30°C. For each time point, cells displaying both the green and the red fluorescence were scored as indicated in the Fig. 1 legend. For each time point, n > 200 (two experiments; error bars show SD). Typical colocalization images corresponding to each type of proteasome localization pattern are shown on the right. Images are maximal projection of z stacks. Bars, 2 μm.
Figure 3.
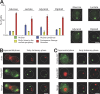
PSGs, actin bodies, and P-bodies are distinct structures. (A) PSGs, as revealed by Pre6p-GFP fluorescence, were observed in cells grown for 4 d in different carbon sources containing rich media and in diploid cells grown in YPDA. For each condition n > 200 (two experiments; error bars show SD). Typical images (maximal projection of z stacks) of cells displaying PSGs (Pre6p-GFP) are shown on the right. (B) Cells expressing Pre6p-GFP were grown in YPDA medium at 30°C, fixed, and stained with Alexa Fluor phalloidin to reveal F-actin–containing structures. Actively proliferating cells (left) displayed actin patches and cables (red) and a typical Pre6p-GFP nuclear localization. Quiescent cells displayed PSGs (green fluorescence) that did not colocalize with actin bodies (red). (C) Cells coexpressing Dcp2p-GFP and Pup1p-RFP were grown in YPDA medium at 30°C. Actively proliferating yeast cells displayed a typical Pup1p-RFP nuclear localization and small and discrete Dcp2p-GFP dots. In quiescent cells, Dcp2p-GFP localized in P-bodies that did not colocalize with PSGs. Images in B and C are single focal planes. Bars, 2 μm.
Figure 4.
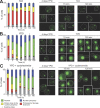
PSGs are rapidly mobilized upon exit from quiescence. Cells expressing Pre6p-GFP were grown for 4 d in YPD medium at 30°C, washed, and either resuspended in water (A) or new YPD medium (B) or preincubated for 1 h in old YPD medium containing 100 μg/ml cycloheximide and then released into new YPD medium containing 100 μg/ml cycloheximide (C). For each time point, cells displaying Pre6p-GFP fluorescence within PSGs, immobile dots close to the nuclear periphery, nuclear periphery, or nucleus were counted (n > 200; two experiments; error bars show SD). The budding index is indicated (n > 200; two experiments; SD < 5%). Images are maximal projection of z stacks. Bar, 2 μm. Of note, the levels of gray for images where PSGs are detected range from 0 to 1,500, and they range from 0 to 500 for images where the fluorescence is detected at the nuclear periphery or in the nucleus.
Figure 5.
S. pombe cells display PSG.S. pombe expressing Pad1-GFP (Rpn11 homologue) were grown for 4 d in YPDA medium at 30°C, washed, and resuspended either in water (A) or new YPDA medium (B) or preincubated for 1 h in old YPDA medium containing 100 μg/ml cycloheximide and then released into new YPDA medium containing 100 μg/ml cycloheximide (C). For each time point, cells displaying Pad1-GFP fluorescence within PSG or at the nuclear periphery were counted (n > 200; two experiments; error bars show SD). Images are maximal projection of z stacks. Bar, 2 μm. Of note, cycloheximide was active on S. pombe cells because 2 h after refeeding in the presence of the drug, cells did not grow and displayed a round-shape morphology, whereas the untreated cells elongated and started to divide. The mean cell size before exit from quiescence was 7.44 ± 0.86 μm (n = 50). 3 h after transfer of quiescent cells to new YPDA medium, mean cell size increased (13 ± 2 μm; n = 50). In contrast, 3 h after transfer of quiescent cells to new YPDA medium containing 100 μg/ml cycloheximide the cell size did not significantly change (8.35 ± 1.16 μm; n = 50).
Similar articles
- Ubiquitin orchestrates proteasome dynamics between proliferation and quiescence in yeast.
Gu ZC, Wu E, Sailer C, Jando J, Styles E, Eisenkolb I, Kuschel M, Bitschar K, Wang X, Huang L, Vissa A, Yip CM, Yedidi RS, Friesen H, Enenkel C. Gu ZC, et al. Mol Biol Cell. 2017 Sep 15;28(19):2479-2491. doi: 10.1091/mbc.E17-03-0162. Epub 2017 Aug 2. Mol Biol Cell. 2017. PMID: 28768827 Free PMC article. - Integrity of the Saccharomyces cerevisiae Rpn11 protein is critical for formation of proteasome storage granules (PSG) and survival in stationary phase.
Saunier R, Esposito M, Dassa EP, Delahodde A. Saunier R, et al. PLoS One. 2013 Aug 6;8(8):e70357. doi: 10.1371/journal.pone.0070357. Print 2013. PLoS One. 2013. PMID: 23936414 Free PMC article. - Proteasome dynamics between proliferation and quiescence stages of Saccharomyces cerevisiae.
Yedidi RS, Fatehi AK, Enenkel C. Yedidi RS, et al. Crit Rev Biochem Mol Biol. 2016 Nov/Dec;51(6):497-512. doi: 10.1080/10409238.2016.1230087. Epub 2016 Sep 28. Crit Rev Biochem Mol Biol. 2016. PMID: 27677933 Review. - Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome.
Pack CG, Yukii H, Toh-e A, Kudo T, Tsuchiya H, Kaiho A, Sakata E, Murata S, Yokosawa H, Sako Y, Baumeister W, Tanaka K, Saeki Y. Pack CG, et al. Nat Commun. 2014 Mar 6;5:3396. doi: 10.1038/ncomms4396. Nat Commun. 2014. PMID: 24598877 - The 26S proteasome of the fission yeast Schizosaccharomyces pombe.
Wilkinson CR, Penney M, McGurk G, Wallace M, Gordon C. Wilkinson CR, et al. Philos Trans R Soc Lond B Biol Sci. 1999 Sep 29;354(1389):1523-32. doi: 10.1098/rstb.1999.0496. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10582238 Free PMC article. Review.
Cited by
- Structure of the reduced microsporidian proteasome bound by PI31-like peptides in dormant spores.
Jespersen N, Ehrenbolger K, Winiger RR, Svedberg D, Vossbrinck CR, Barandun J. Jespersen N, et al. Nat Commun. 2022 Nov 15;13(1):6962. doi: 10.1038/s41467-022-34691-x. Nat Commun. 2022. PMID: 36379934 Free PMC article. - Inactive Proteasomes Routed to Autophagic Turnover Are Confined within the Soluble Fraction of the Cell.
Friedman K, Karmon O, Fridman U, Goldberg Y, Pines O, Ben-Aroya S. Friedman K, et al. Biomolecules. 2022 Dec 30;13(1):77. doi: 10.3390/biom13010077. Biomolecules. 2022. PMID: 36671462 Free PMC article. - Tumor-targeting cell-penetrating peptide, p28, for glioblastoma imaging and therapy.
Mander S, Naffouje SA, Gao J, Li W, Christov K, Green A, Bongarzone ER, Das Gupta TK, Yamada T. Mander S, et al. Front Oncol. 2022 Jul 22;12:940001. doi: 10.3389/fonc.2022.940001. eCollection 2022. Front Oncol. 2022. PMID: 35936749 Free PMC article. - Blm10 facilitates nuclear import of proteasome core particles.
Weberruss MH, Savulescu AF, Jando J, Bissinger T, Harel A, Glickman MH, Enenkel C. Weberruss MH, et al. EMBO J. 2013 Oct 16;32(20):2697-707. doi: 10.1038/emboj.2013.192. Epub 2013 Aug 27. EMBO J. 2013. PMID: 23982732 Free PMC article. - Prolonged starvation drives reversible sequestration of lipid biosynthetic enzymes and organelle reorganization in Saccharomyces cerevisiae.
Suresh HG, da Silveira Dos Santos AX, Kukulski W, Tyedmers J, Riezman H, Bukau B, Mogk A. Suresh HG, et al. Mol Biol Cell. 2015 May 1;26(9):1601-15. doi: 10.1091/mbc.E14-11-1559. Epub 2015 Mar 11. Mol Biol Cell. 2015. PMID: 25761633 Free PMC article.
References
- Bajorek, M., D. Finley, and M.H. Glickman. 2003. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr. Biol. 13:1140–1144. - PubMed
- Collins, G.A., and W.P. Tansey. 2006. The proteasome: a utility tool for transcription? Curr. Opin. Genet. Dev. 16:197–202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases