Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells - PubMed (original) (raw)
Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells
D Bhaumik et al. Oncogene. 2008.
Abstract
Cancer cells often acquire a constitutively active nuclear factor-kappaB (NF-kappaB) program to promote survival, proliferation and metastatic potential by mechanisms that remain largely unknown. Extending observations from an immunologic setting, we demonstrate that microRNA-146a and microRNA-146b (miR-146a/b) when expressed in the highly metastatic human breast cancer cell line MDA-MB-231 function to negatively regulate NF-kappaB activity. Lentiviral-mediated expression of miR-146a/b significantly downregulated interleukin (IL)-1 receptor-associated kinase and TNF receptor-associated factor 6, two key adaptor/scaffold proteins in the IL-1 and Toll-like receptor signaling pathway, known to positively regulate NF-kappaB activity. Impaired NF-kappaB activity was evident from reduced phosphorylation of the NF-kappaB inhibitor IkappaBalpha, reduced NF-kappaB DNA-binding activity and suppressed expression of the NF-kappaB target genes IL-8, IL-6 and matrix metalloproteinase-9. Functionally, miR-146a/b-expressing MDA-MB-231 cells showed markedly impaired invasion and migration capacity relative to control cells. These findings implicate miR-146a/b as a negative regulator of constitutive NF-kappaB activity in a breast cancer setting and suggest that modulating miR-146a/b levels has therapeutic potential to suppress breast cancer metastases.
Figures
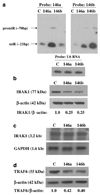
Figure 1. IRAK1 and TRAF6 protein levels are reduced in MDA-MB-231 cells overexpressing miRNA-146a and miR-146b
(a) Northern blot of total RNA prepared from control MDA-MB-231 cells (C), miR-146a-expressing MDA-MB-231 cells (146a) and miR-146b-expressing MDA-MB-231 cells (146b) was probed with an antisense miR-146b DNA oligo (right panel), stripped and reprobed with an antisense miR-146a DNA oligo (left panel). Northern blot was performed as previously described (Scott et al., 2007). Mature and pre-miRNA species are noted. RNA loading was confirmed by probing for the small RNA species, U6 RNA (bottom panel). The lentiviral miR-146a/b expression vectors were constructed by cloning 146 bp of pri miR-146a sequence and 120 bp of pri miR-146b sequence that included the terminal loop, the upper stem and the lower stem with 10 bp of upstream/downstream flanking genomic sequence into a site under cytomegalovirus promoter control. (b) Western blot of total protein lysates prepared from MDA-MB-231 cell pools as described in Figure 1a was probed for IRAK1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The ratio of IRAK1 to β-actin band intensity for each lysate was normalized to the control ratio as shown below the western panel. (c) Northern blot using total RNA isolated from MDA-MB-231 cells as described in (a) was probed for IRAK1. Reprobing the blot for GAPDH established equal RNA loading. The IRAK1 3.2-kb transcript size was deduced from the ethidium-stained gel 18S (1.9 kb) and 28S (5.0 kb) bands. (d) Western blot of total protein lysates as described in (b) was probed for TRAF6 (sc-7221; Santa Cruz Biotechnology). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IRAK1, IL-1 receptor-associated kinase; miRNA, microRNA; TRAF6, tumour-necrosis factor receptor-associated factor 6.
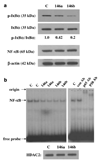
Figure 2. MDA-MB-231 cells overexpressing miR-146a/b have reduced phosphorylation of the NF-κB inhibitor IκBα and reduced NF-κB DNA-binding activity
(a) Western blot of total protein lysates prepared from MDA-MB-231 cell pools as described in Figure 1a was probed for phosphorylated IκBα protein (Ser 32; Cell Signaling Technology, Danvers, MA, USA), stripped and reprobed for total IκBα protein (top panel). The ratio of phosphorylated band intensity to total band intensity was normalized as in Figure 1b and shown below the western panel. Western blot in the lower panel shows that levels of NF-κB (65 kDa) are equal in control cells and miR-146a/b-expressing cells relative to β-actin. (b) EMSA for binding to a consensus NF-κB DNA oligo probe using control (C), miR-146a (146a) and miR-146b (146b) nuclear extracts loaded in duplicate lanes (left panel). The position of the gel origin, NF-κB gel shifted band and free probe are indicated. Right panel shows specificity of NF-κB DNA binding as antibodies to the p65 and p50 subunits of NF-κB supershifted the NF-κB band (p65 Ab and p50 Ab lanes) whereas a control antibody (con Ab, an estrogen receptor antibody) had no effect. Bottom panel: western blot of nuclear extracts used for EMSA probed for HDAC2 (Santa Cruz Biotechnology) shows equal protein content of the extracts. HDAC2, histone deacetylase 2; miR, microRNA; NF-κB, nuclear factor-κB; EMSA, electrophoretic gel shift assays.
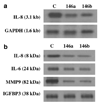
Figure 3. IL-8 mRNA level together with IL-8, IL-6and MMP-9 protein levels are downregulated in cells overexpressing miR-146a/b
(a) Northern blot prepared as described in Figure 1a was probed for IL-8. RNA loading was established by probing the blot with GAPDH. (b) Western blot using TCA-precipitated proteins from conditioned media containing 0.2% serum collected over 24 h was probed for IL-8 (R&D Systems, Minneapolis MN, USA), IL-6 (R&D Systems), MMP-9 (NeoMarkers, Fremont, CA, USA) and IGFBP3 (Santa Cruz Biotechnology). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; miR, microRNA; MMP-9, matrix metalloproteinase 9; TCA, trichloroacetic acid; IGFBP3, insulin like growth factor binding protein-3.
Figure 4. MDA-MB-231 cells overexpressing miR-146a/b have significantly reduced capacity to invade through Matrigel and migrate
(a) Typical field of a Matrigel invasion insert showing the reduced number of invading miR-146a- and miR-146b-expressing cells compared to control cells (8 µm BioCoat Growth Factor Reduced Matrigel Invasion Chambers; BD Biosciences, San Jose, CA, USA). Cells were plated in triplicate at 1 × 105 cells per insert in DMEM media with 10 ng/ml recombinant EGF in DMEM media used as the chemoattractant in the lower chambers for both invasion and migration assays. Following 20 h of incubation, inserts were processed according to the manufacturer’s recommendations using 1% Toluidine Blue. Stained membranes were photographed using an Olympus I × 70 microscope at × 70. b Quantification of invasion achieved by the miR-146a and miR-146b cells as a percentage of that achieved by control cells. Cell counts were determined from the average of five random fields (each field capturing approximately 6% of total membrane area). (c) Quantification of migration through 8-µm pore inserts (BD Biosciences) by miR-146a- and miR-146b-expressing cells as a percentage of that achieved by control cells. All cell-invasion and migration assays were repeated in three independent experiments using the lentiviral-infected miR-146a, miR-146b and control cell pools. DMEM, Dulbecco’s modified Eagle’s medium; miR, microRNA; EGF, epidermal growth factor.
References
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. - PubMed
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Benoy IH, Salgado R, Van Dam P, Geboers K, Van Marck E, Scharpe S, et al. Increased serum interleukin-8 in patients with early and metastatic breast cancer correlates with early dissemination and survival. Clin Cancer Res. 2004;10:7157–7162. - PubMed
- Cao Z, Henzel WJ, Gao X. IRAK: a kinase associated with the interleukin-1 receptor. Science. 1996a;271:1128–1131. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AG025901/AG/NIA NIH HHS/United States
- R01 CA036773/CA/NCI NIH HHS/United States
- R37 AG009909/AG/NIA NIH HHS/United States
- P01-AG025901/AG/NIA NIH HHS/United States
- P50 CA058207/CA/NCI NIH HHS/United States
- R01-CA36773/CA/NCI NIH HHS/United States
- P50-CA58207/CA/NCI NIH HHS/United States
- R37-AG09909/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous