MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor - PubMed (original) (raw)
MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor
Silin Zhong et al. Plant Cell. 2008 May.
Abstract
N6-Methyladenosine is a ubiquitous modification identified in the mRNA of numerous eukaryotes, where it is present within both coding and noncoding regions. However, this base modification does not alter the coding capacity, and its biological significance remains unclear. We show that Arabidopsis thaliana mRNA contains N6-methyladenosine at levels similar to those previously reported for animal cells. We further show that inactivation of the Arabidopsis ortholog of the yeast and human mRNA adenosine methylase (MTA) results in failure of the developing embryo to progress past the globular stage. We also demonstrate that the arrested seeds are deficient in mRNAs containing N6-methyladenosine. Expression of MTA is strongly associated with dividing tissues, particularly reproductive organs, shoot meristems, and emerging lateral roots. Finally, we show that MTA interacts in vitro and in vivo with At FIP37, a homolog of the Drosophila protein FEMALE LETHAL2D and of human WILMS' TUMOUR1-ASSOCIATING PROTEIN. The results reported here provide direct evidence for an essential function for N6-methyladenosine in a multicellular eukaryote, and the interaction with At FIP37 suggests possible RNA processing events that might be regulated or altered by this base modification.
Figures
Figure 1.
Two-dimensional TLC Detection of m6A in Arabidopsis Poly(A) RNA. (A) Two-dimensional TLC analysis of in vitro transcribed RNA containing m6A and normal adenosine. (B) Schematic diagram of the relative positions of nucleotide spots. (C) Two-dimensional TLC analysis of total RNA extracted from 2-week-old Arabidopsis seedlings. (D) Two-dimensional TLC analysis of poly(A) RNA from 2-week-old Arabidopsis seedlings. The m6A:A ratio is 1.5%.
Figure 2.
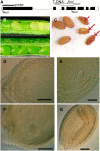
Seeds of the SALK_074069 Insertion in MTA Arrest at the Globular Stage. (A) Arrangement of At4g10760 showing the location of the SALK_074069 T-DNA insertion in exon 4. Exons are shown in black, and introns are shown in white. (B) Siliques from a plant hemizygous for SALK_074069 (top) and from a wild-type control plant (bottom). Seeds homozygous for the SALK_074069 insertion appear white and fail to develop normally. (C) White seeds become shrivelled and nonviable at maturity (arrows). (D) Arrested embryo of the hemizygous SALK_074069. (E) A phenotypically normal embryo from the same silique as in (D) that has reached the heart stage. (F) Although the arrested embryo has not developed past the globular stage, some cell division has continued. (G) A phenotypically normal embryo from the same silique as in (F). Bars = 100 μm.
Figure 3.
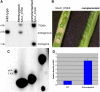
Complementation of SALK_074069 with 35S:MTA. (A) DNA gel blot analysis of wild-type, hemizygous SALK_074069, and homozygous SALK_074069 lines complemented with the MTA cDNA construct. (B) Silique phenotype of plants hemizygous for SALK_074069 (left) and homozygous for both SALK_074069 and the complementing cDNA transgene (right). (C) TLC analysis of the poly(A) RNA purified from the complemented line. (D) qRT-PCR analysis shows a sevenfold higher expression of the MTA transgene in the complemented line. Error bars show
sd
from three replicates.
Figure 4.
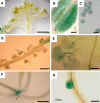
Strong MTA Promoter Activity Is Limited to Discrete Developmental Tissues. GUS expression in various organs of transformed plants is shown. (A) Expression in anthers is initially seen only in tapetal cells (arrow). Bar = 1 cm. (B) Close-up of the staining in (A). Bar = 50 μm. (C) Pollen microspore in a mature anther. Bar = 10 μm. (D) Developing seeds (arrow). Bar = 1 mm. (E) Weak expression is seen in the ovule prior to fertilization (arrow). Bar = 25 μm. (F) In young seedlings, GUS expression is concentrated in the apical meristem (arrow). Bar = 5 mm. (G) In roots, GUS expression is observed primarily during lateral root initiation. Bar = 100 μm.
Figure 5.
m6A Levels in Root, Leaf, and Floral Tissues. (A) Root. (B) Fully expanded leaf. (C) Flower buds. (D) RNA gel blot assay of MTA expression.
Figure 6.
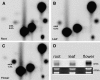
Disruption of MTA Results in the Loss of m6A from the mRNA of Embryo-Defective Seeds. (A) m6A is readily detectable in the mRNA from wild-type seeds. (B) m6A is not detectable in the mRNA from white embryo-defective seeds of SALK_074069, even after prolonged exposure. (C) m6A is readily detectable in the mRNA from white seeds of the control embryo development mutant emb15. (D) RT-PCR showing the absence of MTA transcript in the poly(A) RNA from SALK_074069 white seeds.
Figure 7.
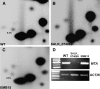
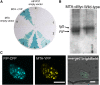
MTA Interacts with FIP37. (A) MTA interacts with FIP37 (b) but does not interact with the empty prey vector, pDEST22 (a). FIP37 does not transactivate GUS expression in the presence of the empty bait vector, pDEST32 (c). ProQuest (Invitrogen) controls for assessing the relative interaction strengths are indicated 1 to 5. (B) Proteins of wild-type and transgenic Arabidopsis plants expressing MTA-c-Myc were extracted under nondenaturing conditions and immunoprecipitated using anti-c-Myc antibody. After SDS-PAGE separation, the anti-FIP37 antibody detected a protein of 48 kD (bottom arrow) in the precipitated extracts of plants expressing the MTA-c-Myc fusion but not in those of the wild-type control. (C) Transient expression of fluorescent protein fusions in onion epidermal peels. FIP37-CFP (left) and MTA-YFP (middle) colocalized to the nucleus. YFP and CFP images were superimposed on the bright-field image to show the position of the nucleus (right). Bars = 10 μm.
Similar articles
- mRNA adenosine methylase (MTA) deposits m6A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana.
Bhat SS, Bielewicz D, Gulanicz T, Bodi Z, Yu X, Anderson SJ, Szewc L, Bajczyk M, Dolata J, Grzelak N, Smolinski DJ, Gregory BD, Fray RG, Jarmolowski A, Szweykowska-Kulinska Z. Bhat SS, et al. Proc Natl Acad Sci U S A. 2020 Sep 1;117(35):21785-21795. doi: 10.1073/pnas.2003733117. Epub 2020 Aug 17. Proc Natl Acad Sci U S A. 2020. PMID: 32817553 Free PMC article. - Functional interdependence of N6-methyladenosine methyltransferase complex subunits in Arabidopsis.
Shen L. Shen L. Plant Cell. 2023 May 29;35(6):1901-1916. doi: 10.1093/plcell/koad070. Plant Cell. 2023. PMID: 36890720 Free PMC article. - N(6)-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis.
Shen L, Liang Z, Gu X, Chen Y, Teo ZW, Hou X, Cai WM, Dedon PC, Liu L, Yu H. Shen L, et al. Dev Cell. 2016 Jul 25;38(2):186-200. doi: 10.1016/j.devcel.2016.06.008. Epub 2016 Jul 7. Dev Cell. 2016. PMID: 27396363 Free PMC article. - The Arabidopsis epitranscriptome.
Fray RG, Simpson GG. Fray RG, et al. Curr Opin Plant Biol. 2015 Oct;27:17-21. doi: 10.1016/j.pbi.2015.05.015. Epub 2015 Jun 3. Curr Opin Plant Biol. 2015. PMID: 26048078 Review. - N6-methyladenosine modification underlies messenger RNA metabolism and plant development.
Shao Y, Wong CE, Shen L, Yu H. Shao Y, et al. Curr Opin Plant Biol. 2021 Oct;63:102047. doi: 10.1016/j.pbi.2021.102047. Epub 2021 May 6. Curr Opin Plant Biol. 2021. PMID: 33965696 Review.
Cited by
- Epigenetic regulations in mammalian spermatogenesis: RNA-m6A modification and beyond.
Gui Y, Yuan S. Gui Y, et al. Cell Mol Life Sci. 2021 Jun;78(11):4893-4905. doi: 10.1007/s00018-021-03823-9. Epub 2021 Apr 9. Cell Mol Life Sci. 2021. PMID: 33835194 Free PMC article. Review. - Transcriptome-Wide Identification of m6A Writers, Erasers and Readers and Their Expression Profiles under Various Biotic and Abiotic Stresses in Pinus massoniana Lamb.
Yao S, Song Y, Cheng X, Wang D, Li Q, Zhang J, Chen Q, Yu Q, Ji K. Yao S, et al. Int J Mol Sci. 2024 Jul 22;25(14):7987. doi: 10.3390/ijms25147987. Int J Mol Sci. 2024. PMID: 39063230 Free PMC article. - N6-methyladenosine RNA modification regulates photosynthesis during photodamage in plants.
Zhang M, Zeng Y, Peng R, Dong J, Lan Y, Duan S, Chang Z, Ren J, Luo G, Liu B, Růžička K, Zhao K, Wang HB, Jin HL. Zhang M, et al. Nat Commun. 2022 Dec 2;13(1):7441. doi: 10.1038/s41467-022-35146-z. Nat Commun. 2022. PMID: 36460653 Free PMC article. - The m6A Dynamics of Profilin in Neurogenesis.
Rockwell AL, Hongay CF. Rockwell AL, et al. Front Genet. 2019 Nov 12;10:987. doi: 10.3389/fgene.2019.00987. eCollection 2019. Front Genet. 2019. PMID: 31798620 Free PMC article. Review. - Progress and application of epitranscriptomic m6A modification in gastric cancer.
Xu Y, Huang C. Xu Y, et al. RNA Biol. 2022 Jan;19(1):885-896. doi: 10.1080/15476286.2022.2096793. RNA Biol. 2022. PMID: 35796515 Free PMC article. Review.
References
- Adams, J.M., and Cory, S. (1975). Modified nucleosides and bizarre 5′-termini in mouse myeloma messenger-RNA. Nature 255 28–33. - PubMed
- Agris, P.F., Vendeixa, F.A.P., and Grahama, W.D. (2007). tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 366 1–13. - PubMed
- Beemon, K., and Keith, J. (1977). Localization of N6-methyladenosine in Rous-sarcoma virus genome. J. Mol. Biol. 113 165–179. - PubMed
- Bjork, G.R., Ericson, J.U., Gustafsson, C.E., Hagervall, T.G., Jonsson, Y.H., and Wikstrom, P.M. (1987). Transfer RNA modification. Annu. Rev. Biochem. 56 263–287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/C513369/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C523369/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous