BatchPrimer3: a high throughput web application for PCR and sequencing primer design - PubMed (original) (raw)
BatchPrimer3: a high throughput web application for PCR and sequencing primer design
Frank M You et al. BMC Bioinformatics. 2008.
Abstract
Background: Microsatellite (simple sequence repeat - SSR) and single nucleotide polymorphism (SNP) markers are two types of important genetic markers useful in genetic mapping and genotyping. Often, large-scale genomic research projects require high-throughput computer-assisted primer design. Numerous such web-based or standard-alone programs for PCR primer design are available but vary in quality and functionality. In particular, most programs lack batch primer design capability. Such a high-throughput software tool for designing SSR flanking primers and SNP genotyping primers is increasingly demanded.
Results: A new web primer design program, BatchPrimer3, is developed based on Primer3. BatchPrimer3 adopted the Primer3 core program as a major primer design engine to choose the best primer pairs. A new score-based primer picking module is incorporated into BatchPrimer3 and used to pick position-restricted primers. BatchPrimer3 v1.0 implements several types of primer designs including generic primers, SSR primers together with SSR detection, and SNP genotyping primers (including single-base extension primers, allele-specific primers, and tetra-primers for tetra-primer ARMS PCR), as well as DNA sequencing primers. DNA sequences in FASTA format can be batch read into the program. The basic information of input sequences, as a reference of parameter setting of primer design, can be obtained by pre-analysis of sequences. The input sequences can be pre-processed and masked to exclude and/or include specific regions, or set targets for different primer design purposes as in Primer3Web and primer3Plus. A tab-delimited or Excel-formatted primer output also greatly facilitates the subsequent primer-ordering process. Thousands of primers, including wheat conserved intron-flanking primers, wheat genome-specific SNP genotyping primers, and Brachypodium SSR flanking primers in several genome projects have been designed using the program and validated in several laboratories.
Conclusion: BatchPrimer3 is a comprehensive web primer design program to develop different types of primers in a high-throughput manner. Additional methods of primer design can be easily integrated into future versions of BatchPrimer3. The program with source code and thousands of PCR and sequencing primers designed for wheat and Brachypodium are accessible at http://wheat.pw.usda.gov/demos/BatchPrimer3/.
Figures
Figure 1
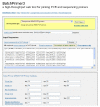
Web interface of BatchPrimer3 v1.0 application. Various types of primer design can be selected from the primer type pull-up combo-box, and corresponding parameter setting panels are placed below the sequence input box. Pre-analysis of input sequences can be performed before batch primer design.
Figure 2
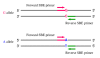
Primer design of single base extension (SBE) primers. One SBE primer is positioned at the base of the 3' end immediately upstream to the SNP.
Figure 3
Primer design of allele-specific (AS) primers. Two AS primers, one for each allele of a SNP are designed. The AS primers contain one of two polymorphic nucleotides at the primer 3' end. Two sets of primers, either forward or reverse primers can be designed. If a common reverse or forward primer is used in a PCR reaction, the reaction is called allele-specific PCR (AS-PCR). Generally, two PCR reactions are needed for detection of both alleles of a SNP. A variant of AS-PCR is to use only one AS primer and two outer SNP-flanking primers in a single PCR reaction, i.e., three-primer nested system [27]. A mismatch (represented by *) may be deliberately introduced at the third position from the 3' end of each of AS primers to increase allelic specificity (See Table 1). The SNP R (G/A) is illustrated as an example, and the other types of SNPs can be applied in the same way.
Figure 4
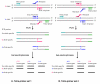
Schematic illustration of primer design for the tetra-primer ARMS PCR (redraw from Ye et al., 2001 [16]). The SNP R (G/A) is presented as an example, and the other types of SNPs can be applied in the same way. Four primers, one pair of inner allele-specific (AS) primers and one pair of outer standard primers, are required in a single PCR reaction. Two AS products, one for the G allele and the other for the A allele are amplified using two pairs of primers. The former consists of a G AS primer and an outer standard primer, and other latter contains an A AS primer and an outer standard primer. A mismatch (represented by *) is deliberately introduced at the third position from the 3' end of each of the two AS primers to increase allelic specificity (See Table 1). Two outer standard primers are designed in such a way that the amplicons of two alleles differ in sizes and can be resolved by agarose gel electrophoresis.
Figure 5
An example of DNA sequence preprocessing. Any unwanted regions for primer design in sequences can be masked using a pair of "<>" to keep the sequence unchanged (A). Alternatively, the unwanted regions can be replaced with "Ns" (B). The included region can be specified by one pair "{}" and only one included region can be masked.
Figure 6
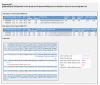
Screenshot of a primer set in batch primer design. The picture shows the primer design results of sequence ID (rs16791736) for tetra-primer ARMS PCR.
Similar articles
- ConservedPrimers 2.0: a high-throughput pipeline for comparative genome referenced intron-flanking PCR primer design and its application in wheat SNP discovery.
You FM, Huo N, Gu YQ, Lazo GR, Dvorak J, Anderson OD. You FM, et al. BMC Bioinformatics. 2009 Oct 13;10:331. doi: 10.1186/1471-2105-10-331. BMC Bioinformatics. 2009. PMID: 19825183 Free PMC article. - Simple sequence repeat marker loci discovery using SSR primer.
Robinson AJ, Love CG, Batley J, Barker G, Edwards D. Robinson AJ, et al. Bioinformatics. 2004 Jun 12;20(9):1475-6. doi: 10.1093/bioinformatics/bth104. Epub 2004 Feb 12. Bioinformatics. 2004. PMID: 14962913 - RExPrimer: an integrated primer designing tool increases PCR effectiveness by avoiding 3' SNP-in-primer and mis-priming from structural variation.
Piriyapongsa J, Ngamphiw C, Assawamakin A, Wangkumhang P, Suwannasri P, Ruangrit U, Agavatpanitch G, Tongsima S. Piriyapongsa J, et al. BMC Genomics. 2009 Dec 3;10 Suppl 3(Suppl 3):S4. doi: 10.1186/1471-2164-10-S3-S4. BMC Genomics. 2009. PMID: 19958502 Free PMC article. - Graphical design of primers with PerlPrimer.
Marshall O. Marshall O. Methods Mol Biol. 2007;402:403-14. doi: 10.1007/978-1-59745-528-2_21. Methods Mol Biol. 2007. PMID: 17951808 Review. - PCR primer design using statistical modeling.
Yuryev A. Yuryev A. Methods Mol Biol. 2007;402:93-104. doi: 10.1007/978-1-59745-528-2_5. Methods Mol Biol. 2007. PMID: 17951792 Review.
Cited by
- De-novo transcriptome assembly for gene identification, analysis, annotation, and molecular marker discovery in Onobrychis viciifolia.
Mora-Ortiz M, Swain MT, Vickers MJ, Hegarty MJ, Kelly R, Smith LM, Skøt L. Mora-Ortiz M, et al. BMC Genomics. 2016 Sep 26;17(1):756. doi: 10.1186/s12864-016-3083-6. BMC Genomics. 2016. PMID: 27671367 Free PMC article. - Rapid identification of causal mutations in tomato EMS populations via mapping-by-sequencing.
Garcia V, Bres C, Just D, Fernandez L, Tai FW, Mauxion JP, Le Paslier MC, Bérard A, Brunel D, Aoki K, Alseekh S, Fernie AR, Fraser PD, Rothan C. Garcia V, et al. Nat Protoc. 2016 Dec;11(12):2401-2418. doi: 10.1038/nprot.2016.143. Epub 2016 Nov 3. Nat Protoc. 2016. PMID: 27809315 - New microsatellite markers for Xerophyta dasylirioides (Velloziaceae), an endemic species on Malagasy inselbergs.
Rexroth J, Krebes L, Wöhrmann T, Harpke D, Rabarimanarivo M, Phillipson P, Weising K, Porembski S. Rexroth J, et al. Appl Plant Sci. 2019 Aug 14;7(8):e11282. doi: 10.1002/aps3.11282. eCollection 2019 Aug. Appl Plant Sci. 2019. PMID: 31467805 Free PMC article. - Assessing introgressive hybridization in roan antelope (Hippotragus equinus): Lessons from South Africa.
van Wyk AM, Dalton DL, Kotzé A, Grobler JP, Mokgokong PS, Kropff AS, Jansen van Vuuren B. van Wyk AM, et al. PLoS One. 2019 Oct 18;14(10):e0213961. doi: 10.1371/journal.pone.0213961. eCollection 2019. PLoS One. 2019. PMID: 31626669 Free PMC article. - ConservedPrimers 2.0: a high-throughput pipeline for comparative genome referenced intron-flanking PCR primer design and its application in wheat SNP discovery.
You FM, Huo N, Gu YQ, Lazo GR, Dvorak J, Anderson OD. You FM, et al. BMC Bioinformatics. 2009 Oct 13;10:331. doi: 10.1186/1471-2105-10-331. BMC Bioinformatics. 2009. PMID: 19825183 Free PMC article.
References
- Abd-Elsalam KA. Bioinformatics tools and guideline for PCR primer design. Africa Journal of Biotechnology. 2003;2:91–95.
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editor. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ , Humana Press; 2000. pp. 365–386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources