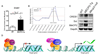MeCP2, a key contributor to neurological disease, activates and represses transcription - PubMed (original) (raw)
MeCP2, a key contributor to neurological disease, activates and represses transcription
Maria Chahrour et al. Science. 2008.
Abstract
Mutations in the gene encoding the transcriptional repressor methyl-CpG binding protein 2 (MeCP2) cause the neurodevelopmental disorder Rett syndrome. Loss of function as well as increased dosage of the MECP2 gene cause a host of neuropsychiatric disorders. To explore the molecular mechanism(s) underlying these disorders, we examined gene expression patterns in the hypothalamus of mice that either lack or overexpress MeCP2. In both models, MeCP2 dysfunction induced changes in the expression levels of thousands of genes, but unexpectedly the majority of genes (approximately 85%) appeared to be activated by MeCP2. We selected six genes and confirmed that MeCP2 binds to their promoters. Furthermore, we showed that MeCP2 associates with the transcriptional activator CREB1 at the promoter of an activated target but not a repressed target. These studies suggest that MeCP2 regulates the expression of a wide range of genes in the hypothalamus and that it can function as both an activator and a repressor of transcription.
Figures
Fig. 1
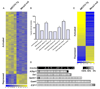
Significant gene expression changes in hypothalami of MeCP2 mouse models. (A) Heatmap showing hypothalamic gene expression profiles in _MECP2_-Tg and _Mecp2_-null mice. Yellow and blue colors indicate increased and decreased expression, respectively, relative to WT. Each column represents one RNA sample from each genotype, and each row represents one gene. Expression levels are depicted according to the color scale at the bottom. 2,184 genes are activated in the presence of MeCP2, while 377 genes are repressed (FDR-adjusted p-value < 0.05). (B) Promoter regions of genes upregulated by MeCP2 contain significantly more CpG islands compared to those of genes downregulated by MeCP2 (* p≤0.006). (C) Validation of expression changes for 66 genes by quantitative real-time RT-PCR analysis. Gene expression levels from the microarray were validated in four _MECP2_-Tg males and four _Mecp2_-null males. Data are plotted as fold up- (yellow) or downregulation (blue) over WT (p<0.05, t test). Each row represents a single gene, and each column represents data for four samples from each genotype. Levels are depicted according to the color scale at the bottom. The complete list of validated genes is available in Supporting Online Material (12) (table S7). (D) Bisulfite sequencing revealed that CpG sites in the promoters of the repressed target genes Grin2a and A2bp1 are heavily methylated, while those of activated targets Sst, Gprin1, Gamt, and E2F1 are not. CpG sites are depicted as squares, with the percent methylation presented according to the scale. The data were generated from three independent animals and for each promoter ~10 clones were sequenced.
Fig. 2
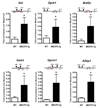
MeCP2 binds to the promoter region of six target genes. ChIP with anti-MeCP2 antibody shows that MeCP2 binds to the promoter regions of activated targets Sst, Oprk1, Gamt, and Gprin1, and repressed targets Mef2c and A2bp1. The red line indicates the location of the probe and primers (relative to the start site (+1), in bps or in kb, k) and the closest CpG islands in the promoter region are depicted. Quantitative real-time PCR values were normalized to the input and plotted as fold enrichment over _Mecp2_-null (N ≥ 3, * p≤0.03, t test).
Fig. 3
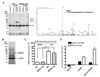
Physical and functional cooperation between MeCP2 and CREB1. (A) Specific interaction between MeCP2 and CREB1 in chromatin. Mouse brain tissue was fractionated into cytosolic, nuclear, and chromatin fractions for protein extraction from WT and _Mecp2_-null mice. Protein extracts were immunoprecipitated with an anti-MeCP2 antibody and resolved by SDS-PAGE. Each lane was divided into eight sections (brackets) and analyzed by mass spectrometry. Proteins identified in the WT but not in the knockout sample are considered specific MeCP2 interacting proteins. MeCP2 was identified only in the chromatin fraction from WT mice (★). A representative nano-HPLC/MS/MS spectrum is shown that identifies CREB1 as one of the specific MeCP2 binding proteins in the 35–50 kDa region in SDS-PAGE indicated by υ. The complete list of interacting proteins is available in Supporting Online Material (12) (table S8). (B) MeCP2 was detected after immunoprecipitation with anti-CREB1 from Neuro2a cells, using mass spectrometry analysis of the indicated band. (C) MeCP2 associates with CREB1 at the promoter of an activated but not a repressed target. SeqChIP analysis detects co-occupancy of MeCP2 and CREB1 at the promoter of the activated MeCP2 target Sst, but not at the promoter of the repressed target Mef2c. The primary ChIP was performed with an anti-MeCP2 antibody and the secondary ChIP was done with an anti-CREB1 antibody. Quantitative real-time PCR values were normalized to the input and plotted as percent of secondary over primary ChIP (N = 3, * p<0.001, two-way ANOVA). (D) Functional synergy between MeCP2 and CREB1 at the promoter of an activated target. Luciferase assay in Neuro2a cells reveals synergistic activation at the promoter of the activated MeCP2 target Sst but not the repressed target A2bp1 (N = 3, * p<0.002, two-way ANOVA).
Fig. 4
Creb1 is a direct MeCP2 target. (A) ChIP analysis revealed binding of MeCP2 to the promoter region of Creb1. Quantitative real-time PCR values were normalized to the input and plotted as fold enrichment over _Mecp2_-null (N = 4, * p<0.05, t test). (B) ChIP analysis using a custom array with probes to the Creb1 promoter. Genomic locations of the array probes are indicated on the X-axis and the dashed vertical line indicates location of the quantitative real-time PCR probe. The red line represents the normalized net signal from the _Mecp2_-null samples. For the WT and _MECP2_-Tg samples, normalized net intensities from each probe are plotted and are significantly different from the _Mecp2_-null near the predicted MeCP2 binding site (* p<0.02). (C) CREB1 and Sst protein levels are increased in _MECP2_-Tg hypothalami compared to WT. Western blot is representative of three animals from each genotype. (D) MeCP2 can function as a transcriptional activator and repressor in the hypothalamus (CpG sites are depicted as blue circles irrespective of their methylation status).
Comment in
- Medicine. Activating a repressor.
Cohen S, Zhou Z, Greenberg ME. Cohen S, et al. Science. 2008 May 30;320(5880):1172-3. doi: 10.1126/science.1159146. Science. 2008. PMID: 18511680 Free PMC article. No abstract available.
Similar articles
- Cerebellar gene expression profiles of mouse models for Rett syndrome reveal novel MeCP2 targets.
Jordan C, Li HH, Kwan HC, Francke U. Jordan C, et al. BMC Med Genet. 2007 Jun 20;8:36. doi: 10.1186/1471-2350-8-36. BMC Med Genet. 2007. PMID: 17584923 Free PMC article. - Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus.
Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Ben-Shachar S, et al. Hum Mol Genet. 2009 Jul 1;18(13):2431-42. doi: 10.1093/hmg/ddp181. Epub 2009 Apr 15. Hum Mol Genet. 2009. PMID: 19369296 Free PMC article. - MeCP2 deficiency in the brain decreases BDNF levels by REST/CoREST-mediated repression and increases TRKB production.
Abuhatzira L, Makedonski K, Kaufman Y, Razin A, Shemer R. Abuhatzira L, et al. Epigenetics. 2007 Oct-Dec;2(4):214-22. doi: 10.4161/epi.2.4.5212. Epub 2007 Oct 29. Epigenetics. 2007. PMID: 18075316 - [Research progress of Rett syndrome causing gene MECP2--the structure, function and modulation of MECP2].
Zhang JJ, Bao XH. Zhang JJ, et al. Beijing Da Xue Xue Bao Yi Xue Ban. 2009 Dec 18;41(6):712-5. Beijing Da Xue Xue Bao Yi Xue Ban. 2009. PMID: 20019788 Review. Chinese. - MeCP2 in Rett syndrome: transcriptional repressor or chromatin architectural protein?
Chadwick LH, Wade PA. Chadwick LH, et al. Curr Opin Genet Dev. 2007 Apr;17(2):121-5. doi: 10.1016/j.gde.2007.02.003. Epub 2007 Feb 20. Curr Opin Genet Dev. 2007. PMID: 17317146 Review.
Cited by
- Dysregulation of the long non-coding RNA transcriptome in a Rett syndrome mouse model.
Petazzi P, Sandoval J, Szczesna K, Jorge OC, Roa L, Sayols S, Gomez A, Huertas D, Esteller M. Petazzi P, et al. RNA Biol. 2013 Jul;10(7):1197-203. doi: 10.4161/rna.24286. Epub 2013 Apr 17. RNA Biol. 2013. PMID: 23611944 Free PMC article. - Prospects for the development of epigenetic drugs for CNS conditions.
Szyf M. Szyf M. Nat Rev Drug Discov. 2015 Jul;14(7):461-74. doi: 10.1038/nrd4580. Epub 2015 May 22. Nat Rev Drug Discov. 2015. PMID: 26000723 Review. - Astrocytes conspire with neurons during progression of neurological disease.
McGann JC, Lioy DT, Mandel G. McGann JC, et al. Curr Opin Neurobiol. 2012 Oct;22(5):850-8. doi: 10.1016/j.conb.2012.03.009. Epub 2012 Apr 3. Curr Opin Neurobiol. 2012. PMID: 22475461 Free PMC article. Review. - Reciprocal control of translation and transcription in autism spectrum disorder.
Longo F, Klann E. Longo F, et al. EMBO Rep. 2021 Jun 4;22(6):e52110. doi: 10.15252/embr.202052110. Epub 2021 May 11. EMBO Rep. 2021. PMID: 33977633 Free PMC article. Review. - MiR-130a regulates neurite outgrowth and dendritic spine density by targeting MeCP2.
Zhang Y, Chen M, Qiu Z, Hu K, McGee W, Chen X, Liu J, Zhu L, Wu JY. Zhang Y, et al. Protein Cell. 2016 Jul;7(7):489-500. doi: 10.1007/s13238-016-0272-7. Epub 2016 Jun 1. Protein Cell. 2016. PMID: 27245166 Free PMC article.
References
- Amir RE, et al. Nat Genet. 1999;23:185. - PubMed
- Moretti P, Zoghbi HY. Curr Opin Genet Dev. 2006;16:276. - PubMed
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. Nat Genet. 2001;27:322. - PubMed
- Collins AL, et al. Hum Mol Genet. 2004;13:2679. - PubMed
- Nan X, et al. Nature. 1998;393:386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS057819/NS/NINDS NIH HHS/United States
- HD024064/HD/NICHD NIH HHS/United States
- R01 NS057819/NS/NINDS NIH HHS/United States
- R01 NS057819-02/NS/NINDS NIH HHS/United States
- P30 HD024064/HD/NICHD NIH HHS/United States
- P30 HD024064-19/HD/NICHD NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases