Roles of RNA polymerase IV in gene silencing - PubMed (original) (raw)
Review
Roles of RNA polymerase IV in gene silencing
Craig S Pikaard et al. Trends Plant Sci. 2008 Jul.
Abstract
Eukaryotes typically have three multi-subunit enzymes that decode the nuclear genome into RNA: DNA-dependent RNA polymerases I, II and III (Pol I, II and III). Remarkably, higher plants have five multi-subunit nuclear RNA polymerases: the ubiquitous Pol I, II and III, which are essential for viability; plus two non-essential polymerases, Pol IVa and Pol IVb, which specialize in small RNA-mediated gene silencing pathways. There are numerous examples of phenomena that require Pol IVa and/or Pol IVb, including RNA-directed DNA methylation of endogenous repetitive elements, silencing of transgenes, regulation of flowering-time genes, inducible regulation of adjacent gene pairs, and spreading of mobile silencing signals. Although biochemical details concerning Pol IV enzymatic activities are lacking, genetic evidence suggests several alternative models for how Pol IV might function.
Figures
Figure 1
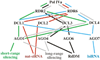
Catalytic subunits of DNA-dependent RNA polymerases (a) The largest and second-largest subunits form the catalytic center. The image is a surface rendering generated using the crystal coordinates for a yeast Pol II elongation complex determined by Westover, Bushnell and Kornberg (PDB:1R9T). Only the two largest Pol II subunits are shown. The DNA template strand is shown in blue, the non-template strand in green and the nascent RNA in red. (b) Domain structures of RNAP largest subunits. E.coli (Ec RPOC) and yeast Pol II largest subunits (Sc RPB1) are compared to the Arabidopsis largest subunits for Pol I (At NRPA1), Pol II (At NRPB1), Pol III (At NRPC1), Pol IVa (At NRPD1a) and Pol IVb (At NRPD1b). Positions of conserved domains A–H are highlighted. Numbers below Pol IV domains are % identities to corresponding Arabidopsis Pol II subunit domains. CTDs of yeast and Arabidopsis Pol II largest subunits have 26 or 39 copies, respectively, of a seven amino acid (heptad) repeat. The domain with similarity to the Defective Chloroplasts and Leaves gene (DeCL domain), present in the CTDs of the Pol IVa and Pol IVb largest subunits, is shown in green. The CTD of NRPD1b also includes a region rich in WG/GW motifs, overlapping ten imperfect 16 amino acid repeats, and a domain composed of alternating glutamines and serines (QS-rich domain). (c) Domain structures of RNAP second-largest subunits. E.coli (Ec RPOB) and yeast Pol II subunits (Sc RPB2) are compared to the Arabidopsis second-largest subunits for Pol I (At NRPA2) , Pol II (At NRPB2), Pol III (At NRPC2) or Pol IV (At NRPD2). Positions of conserved domains A–I are highlighted. Numbers below Pol IV domains are % identities to the corresponding Arabidopsis Pol II subunit domains.
Figure 2
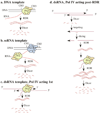
A variety of proteins participate in Pol IVa-dependent silencing pathways. The figure shows a subset of the proteins that are involved in RdDM, nat-siRNA, lsiRNA, short-range silencing and long-distance silencing pathways. Proteins involved in the various pathways are linked by color-coded lines. The diagram does not imply the order of events, but illustrates the diversity of functional collaborations that are possible. Not all mutants have been tested in every pathway, therefore other potential connections may exist. However, the figure reflects the models provided by the authors of the studies discussed in the text.
Figure 3
Possible modes of Pol IVa function. Pol IVa may transcribe a specialized DNA template, such as methylated DNA (a) or single-stranded RNA transcripts derived from methylated DNA loci (b). Alternatively, Pol IVa may transcribe dsRNA generated from bidirectional transcripts, including transcripts of natural antisense gene pairs, or dsRNAs resulting from the annealing of long-distance mobile RNAs with target mRNAs (c and d). The model shown in D may account for the involvement of multiple dicers and multiple RDR inputs in the nat-siRNA and long-distance silencing pathways.
Similar articles
- Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing.
Haag JR, Pontes O, Pikaard CS. Haag JR, et al. PLoS One. 2009;4(1):e4110. doi: 10.1371/journal.pone.0004110. Epub 2009 Jan 1. PLoS One. 2009. PMID: 19119310 Free PMC article. - Ancient Origin and Recent Innovations of RNA Polymerase IV and V.
Huang Y, Kendall T, Forsythe ES, Dorantes-Acosta A, Li S, Caballero-Pérez J, Chen X, Arteaga-Vázquez M, Beilstein MA, Mosher RA. Huang Y, et al. Mol Biol Evol. 2015 Jul;32(7):1788-99. doi: 10.1093/molbev/msv060. Epub 2015 Mar 12. Mol Biol Evol. 2015. PMID: 25767205 Free PMC article. - RNA Pol IV and V in gene silencing: Rebel polymerases evolving away from Pol II's rules.
Zhou M, Law JA. Zhou M, et al. Curr Opin Plant Biol. 2015 Oct;27:154-64. doi: 10.1016/j.pbi.2015.07.005. Epub 2015 Sep 5. Curr Opin Plant Biol. 2015. PMID: 26344361 Free PMC article. Review. - Diverse gene-silencing mechanisms with distinct requirements for RNA polymerase subunits in Zea mays.
Sloan AE, Sidorenko L, McGinnis KM. Sloan AE, et al. Genetics. 2014 Nov;198(3):1031-42. doi: 10.1534/genetics.114.168518. Epub 2014 Aug 27. Genetics. 2014. PMID: 25164883 Free PMC article. - RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants.
Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J, Matzke AJ, Matzke M. Huettel B, et al. Biochim Biophys Acta. 2007 May-Jun;1769(5-6):358-74. doi: 10.1016/j.bbaexp.2007.03.001. Epub 2007 Mar 12. Biochim Biophys Acta. 2007. PMID: 17449119 Review.
Cited by
- Transgenic epigenetics: using transgenic organisms to examine epigenetic phenomena.
McEachern LA. McEachern LA. Genet Res Int. 2012;2012:689819. doi: 10.1155/2012/689819. Epub 2012 Mar 27. Genet Res Int. 2012. PMID: 22567397 Free PMC article. - Re-sequencing and transcriptome analysis reveal rich DNA variations and differential expressions of fertility-related genes in neo-tetraploid rice.
Bei X, Shahid MQ, Wu J, Chen Z, Wang L, Liu X. Bei X, et al. PLoS One. 2019 Apr 5;14(4):e0214953. doi: 10.1371/journal.pone.0214953. eCollection 2019. PLoS One. 2019. PMID: 30951558 Free PMC article. - Transgene expression and transgene-induced silencing in diploid and autotetraploid Arabidopsis.
Finn TE, Wang L, Smolilo D, Smith NA, White R, Chaudhury A, Dennis ES, Wang MB. Finn TE, et al. Genetics. 2011 Feb;187(2):409-23. doi: 10.1534/genetics.110.124370. Epub 2010 Nov 15. Genetics. 2011. PMID: 21078688 Free PMC article. - RNA interference and heterochromatin assembly.
Volpe T, Martienssen RA. Volpe T, et al. Cold Spring Harb Perspect Biol. 2011 Sep 1;3(9):a003731. doi: 10.1101/cshperspect.a003731. Cold Spring Harb Perspect Biol. 2011. PMID: 21441597 Free PMC article. Review. - Regulation of de novo-initiated RNA synthesis in hepatitis C virus RNA-dependent RNA polymerase by intermolecular interactions.
Chinnaswamy S, Murali A, Li P, Fujisaki K, Kao CC. Chinnaswamy S, et al. J Virol. 2010 Jun;84(12):5923-35. doi: 10.1128/JVI.02446-09. Epub 2010 Apr 7. J Virol. 2010. PMID: 20375156 Free PMC article.
References
- Werner F. Structure and function of archaeal RNA polymerases. Mol Microbiol. 2007;65:1395–1404. - PubMed
- Kettenberger H, et al. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. - PubMed
- The-Arabidopsis-Genome-Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
- Onodera Y, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. - PubMed
- Herr AJ, et al. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM060380/GM/NIGMS NIH HHS/United States
- R01 GM077590/GM/NIGMS NIH HHS/United States
- R01GM60380/GM/NIGMS NIH HHS/United States
- R01GM077590/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources