Positive and negative selection on noncoding DNA in Drosophila simulans - PubMed (original) (raw)
Positive and negative selection on noncoding DNA in Drosophila simulans
Penelope R Haddrill et al. Mol Biol Evol. 2008 Sep.
Abstract
There is now a wealth of evidence that some of the most important regions of the genome are found outside those that encode proteins, and noncoding regions of the genome have been shown to be subject to substantial levels of selective constraint, particularly in Drosophila. Recent work has suggested that these regions may also have been subject to the action of positive selection, with large fractions of noncoding divergence having been driven to fixation by adaptive evolution. However, this work has focused on Drosophila melanogaster, which is thought to have experienced a reduction in effective population size (N(e)), and thus a reduction in the efficacy of selection, compared with its closest relative Drosophila simulans. Here, we examine patterns of evolution at several classes of noncoding DNA in D. simulans and find that all noncoding DNA is subject to the action of negative selection, indicated by reduced levels of polymorphism and divergence and a skew in the frequency spectrum toward rare variants. We find that the signature of negative selection on noncoding DNA and nonsynonymous sites is obscured to some extent by purifying selection acting on preferred to unpreferred synonymous codon mutations. We investigate the extent to which divergence in noncoding DNA is inferred to be the product of positive selection and to what extent these inferences depend on selection on synonymous sites and demography. Based on patterns of polymorphism and divergence for different classes of synonymous substitution, we find the divergence excess inferred in noncoding DNA and nonsynonymous sites in the D. simulans lineage difficult to reconcile with demographic explanations.
Figures
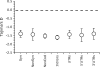
FIG. 1.—
Mean Tajima's D values for coding and noncoding DNA in Drosophila simulans, using the Drosophila melanogaster outgroup. The dashed line indicates the mean expected value of Tajima's D under a model of neutral evolution for a sample of 20 individuals with similar levels of variability to those found here (no recombination, see table 1 of Tajima [1989]). Error bars indicate 2 standard errors. Syn: synonymous sites, NonSyn: nonsynonymous sites, and NonCod: pooled noncoding sites.
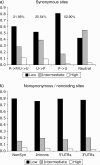
FIG. 2.—
The distribution of frequency classes of polymorphisms for different types of synonymous site changes, nonsynonymous site changes, and noncoding DNA changes. Within synonymous site changes, P → P/U → U includes preferred to preferred and unpreferred to unpreferred changes, P → U are preferred to unpreferred changes, and U → P are unpreferred to preferred changes. “Neutral” indicates the neutral expectation. The low-frequency class includes polymorphisms at a frequency of 1–2/20, the intermediate frequency class 3–17/20, and the high-frequency class 18–19/20. The numbers above each type of synonymous site change indicate the percentage of the total synonymous polymorphisms of each type.
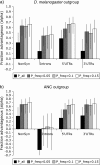
FIG. 3.—
Estimates of the fraction of adaptively driven nucleotide substitutions (α) in coding and noncoding DNA (a) between Drosophila simulans and Drosophila melanogaster and (b) inferred along the D. simulans lineage using a reconstructed ancestor. NonSyn: nonsynonymous sites. For each class of sequence, 4 estimates are presented based on the subset of polymorphisms that were included in the analysis: P_all, all polymorphisms included; P_freq > 0.05, polymorphisms at a frequency of greater than 5% (singletons) included; P_freq > 0.1, polymorphisms at a frequency of greater than 10% (doubletons) included; and P_freq > 0.15, polymorphisms at a frequency of greater than 15% (tripletons) included. Error bars indicate 90% CIs.
Similar articles
- Inferring weak selection from patterns of polymorphism and divergence at "silent" sites in Drosophila DNA.
Akashi H. Akashi H. Genetics. 1995 Feb;139(2):1067-76. doi: 10.1093/genetics/139.2.1067. Genetics. 1995. PMID: 7713409 Free PMC article. - Adaptive evolution of non-coding DNA in Drosophila.
Andolfatto P. Andolfatto P. Nature. 2005 Oct 20;437(7062):1149-52. doi: 10.1038/nature04107. Nature. 2005. PMID: 16237443 - DNA variability and divergence at the notch locus in Drosophila melanogaster and D. simulans: a case of accelerated synonymous site divergence.
DuMont VB, Fay JC, Calabrese PP, Aquadro CF. DuMont VB, et al. Genetics. 2004 May;167(1):171-85. doi: 10.1534/genetics.167.1.171. Genetics. 2004. PMID: 15166145 Free PMC article. - Patterns of polymorphism and divergence from noncoding sequences of Drosophila melanogaster and D. simulans: evidence for nonequilibrium processes.
Kern AD, Begun DJ. Kern AD, et al. Mol Biol Evol. 2005 Jan;22(1):51-62. doi: 10.1093/molbev/msh269. Epub 2004 Sep 29. Mol Biol Evol. 2005. PMID: 15456897 Review. - Codon bias evolution in Drosophila. Population genetics of mutation-selection drift.
Akashi H. Akashi H. Gene. 1997 Dec 31;205(1-2):269-78. doi: 10.1016/s0378-1119(97)00400-9. Gene. 1997. PMID: 9461401 Review.
Cited by
- Drosophila functional elements are embedded in structurally constrained sequences.
Kenigsberg E, Tanay A. Kenigsberg E, et al. PLoS Genet. 2013 May;9(5):e1003512. doi: 10.1371/journal.pgen.1003512. Epub 2013 May 30. PLoS Genet. 2013. PMID: 23750124 Free PMC article. - Evolutionary genomics of Colias Phosphoglucose Isomerase (PGI) introns.
Wang B, Mason Depasse J, Watt WB. Wang B, et al. J Mol Evol. 2012 Feb;74(1-2):96-111. doi: 10.1007/s00239-012-9492-5. Epub 2012 Mar 3. J Mol Evol. 2012. PMID: 22388354 - Population history in Arabidopsis halleri using multilocus analysis.
Heidel AJ, Ramos-Onsins SE, Wang WK, Chiang TY, Mitchell-Olds T. Heidel AJ, et al. Mol Ecol. 2010 Aug;19(16):3364-79. doi: 10.1111/j.1365-294X.2010.04761.x. Epub 2010 Jul 28. Mol Ecol. 2010. PMID: 20670364 Free PMC article. - Does positive selection drive transcription factor binding site turnover? A test with Drosophila cis-regulatory modules.
He BZ, Holloway AK, Maerkl SJ, Kreitman M. He BZ, et al. PLoS Genet. 2011 Apr;7(4):e1002053. doi: 10.1371/journal.pgen.1002053. Epub 2011 Apr 28. PLoS Genet. 2011. PMID: 21572512 Free PMC article. - Selection on codon usage and base composition in Drosophila americana.
de Procé SM, Zeng K, Betancourt AJ, Charlesworth B. de Procé SM, et al. Biol Lett. 2012 Feb 23;8(1):82-5. doi: 10.1098/rsbl.2011.0601. Epub 2011 Aug 17. Biol Lett. 2012. PMID: 21849309 Free PMC article.
References
- Akashi H. Detecting the ‘footprint’ of natural selection in within and between species DNA sequence data. Gene. 1999;238:39–51. - PubMed
- Andolfatto P. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol Biol Evol. 2001;18:279–290. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases