Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex - PubMed (original) (raw)
. 2008 Jul;36(12):3993-4008.
doi: 10.1093/nar/gkn349. Epub 2008 May 31.
Affiliations
- PMID: 18515835
- PMCID: PMC2475620
- DOI: 10.1093/nar/gkn349
Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex
Henri-Marc Bourbon. Nucleic Acids Res. 2008 Jul.
Abstract
The multisubunit Mediator (MED) complex bridges DNA-bound transcriptional regulators to the RNA polymerase II (PolII) initiation machinery. In yeast, the 25 MED subunits are distributed within three core subcomplexes and a separable kinase module composed of Med12, Med13 and the Cdk8-CycC pair thought to control the reversible interaction between MED and PolII by phosphorylating repeated heptapeptides within the Rpb1 carboxyl-terminal domain (CTD). Here, MED conservation has been investigated across the eukaryotic kingdom. Saccharomyces cerevisiae Med2, Med3/Pgd1 and Med5/Nut1 subunits are apparent homologs of metazoan Med29/Intersex, Med27/Crsp34 and Med24/Trap100, respectively, and these and other 30 identified human MED subunits have detectable counterparts in the amoeba Dictyostelium discoideum, indicating that none is specific to metazoans. Indeed, animal/fungal subunits are also conserved in plants, green and red algae, entamoebids, oomycetes, diatoms, apicomplexans, ciliates and the 'deep-branching' protists Trichomonas vaginalis and Giardia lamblia. Surprisingly, although lacking CTD heptads, T. vaginalis displays 44 MED subunit homologs, including several CycC, Med12 and Med13 paralogs. Such observations have allowed the identification of a conserved 17-subunit framework around which peripheral subunits may be assembled, and support a very ancient eukaryotic origin for a large, four-module MED. The implications of this comprehensive work for MED structure-function relationships are discussed.
Figures
Figure 1.
Conserved motifs and domains among fungal and human MED subunits. (A) Architectural organization of S. cerevisiae MED. Subunits have been assigned to Tail (yellow), Middle (green), Head (blue) and Cdk8 kinase (red) modules according to an integrated interaction map from ref. (25), those essential for cell viability being depicted by darker colours. (B–C) Distribution of conserved SSMs within the primary sequences of the 25 identified S. cerevisiae MED subunits (A) and of the eight human MED subunits found thus far only in animals (B). SSMs are represented by black boxes, except those (for Cdk8 and CycC) located within broadly conserved kinase and cyclin repeat domains (in black), which are depicted by white boxes. As indicated, for each subunit, they have been numbered from the N- toward the C-terminus. For Med15, Med16, Med25, Med26 and Med27 the extents of the distinguishable domains found in many functionally unrelated proteins are shown above the corresponding SSMs. Of note, for most subunits the SSMs are distributed throughout the primary sequences. Drawings of fungal Med2, Med3 and Med5 subunits and those of their predicted human counterparts Med29, Med27 and Med24, respectively, are included within boxes coloured according to the novel orthology assignments shown in Figure 2.
Figure 2.
Fungal Med2, Med3 and Med5 are apparent homologs of metazoan Med29, Med27 and Med24, respectively. (A–C) PSI-blast alignments of human (Hs) Med29 (A), Med27 (B) and Med24 (C) with S. cerevisiae (Sc) Med2, Med3 and Med24 primary sequences, respectively, are shown. Numbers indicate amino-acid positions within human (top lines) and fungal (bottom lines) primary sequences. Portions corresponding to evolutionarily conserved SSMs are denoted by dashed-line boxes.
Figure 2.
Fungal Med2, Med3 and Med5 are apparent homologs of metazoan Med29, Med27 and Med24, respectively. (A–C) PSI-blast alignments of human (Hs) Med29 (A), Med27 (B) and Med24 (C) with S. cerevisiae (Sc) Med2, Med3 and Med24 primary sequences, respectively, are shown. Numbers indicate amino-acid positions within human (top lines) and fungal (bottom lines) primary sequences. Portions corresponding to evolutionarily conserved SSMs are denoted by dashed-line boxes.
Figure 3.
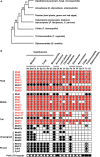
Mediator subunits conserved across the eukaryotic kingdom. (A) Schematic representation of the best tree from amino-acid sequence analyses of 22 evolutionarily conserved protein and two rRNA genes collected from species distributed among eight higher-order groups of eukaryotes. Bootstrap proportion (BP) values are shown on internal branches [after ref. (106)]. (B) Subunits are grouped to Head, Middle, Tail and Cdk8 kinase modules according to the current model of S. cerevisiae MED architecture (25), those underlined being critically required for cell viability (Figure 1A). Except possibly Med23 (see text), the subunits only identified to date in purified metazoan MED complexes remain to be assigned to a specific module. Indicated species are representatives of the examined eukaryotic taxa (for a comprehensive analysis see Figure S6): H. sapiens (Hs), D. melanogaster (Dm), C. elegans (Ce), S. cerevisiae (Sc), S. pombe (Sp), C. cinereus (Cc), R. oryzae (Ro), E. cuniculi (Ec), D. discoideum (Dd), E. histolytica (Eh), A. thaliana (At), C. reinhardtii (Cr), G. sulphuraria (Gs), P. sojae (Ps), T. pseudonana (Th), P. falciparum (Pf), C. parvum (Cp), T. thermophila (Tt), T. vaginalis (Tv) and G. lamblia (Gl). Except for Med28 and Med29/Med2 for which no conserved domain signatures have been yet defined, HHpred analyses predicted with high statistical confidence (Prob >98%, with _E_-values >1.0e-7) that most if not all identified proteins are likely bona fide MED subunits (see dataset S8). Dots indicate that at least one apparent homolog is present in the investigated genomes. In the cases where several paralogs were detected, their number is indicated. Broadly conserved subunits, which include those essential for yeast viability, are depicted in red and may constitute a framework around which other subunits are assembled. White dots in the bottom row indicate the presence of repetitive heptads within the Rpb1 CTD.
Figure 4.
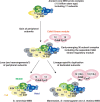
A model for MED architectural organization during eukaryotic evolution, from a primitive scaffold to current complexes. MED subunits have been assigned to Head, Middle, Tail and Cdk8 kinase modules according to a connection map from S. cerevisiae (25), as shown in Figure 1A. Subunits initially identified in purified metazoan complexes and without homologs in budding yeast MED are indicated in grey. Hypothetical structural organizations of the present-day mammalian, Drosophila and Arabidopsis MED complexes as inferred from the current yeast model, orthology assignments and other functional studies. Broken circles outline that Med1 and Med26 subunits are apparently absent in plants (98). Structural rearrangements, notably the location of the Med2/29–Med3/27–Med15 subunits (see text), may have occurred during evolution. Note that subunits of the separable Cdk8 module, which may also have appeared early on during eukaryotic evolution, have detectable homologs (and in a few cases paralogs) only in some extant taxa. It is also noteworthy that during metazoan diversification Med1, Med12 and Med13 have considerably increased their size and have been duplicated in some (sub)-phylum.
Similar articles
- Distinct roles for Mediator Cdk8 module subunits in Drosophila development.
Loncle N, Boube M, Joulia L, Boschiero C, Werner M, Cribbs DL, Bourbon HM. Loncle N, et al. EMBO J. 2007 Feb 21;26(4):1045-54. doi: 10.1038/sj.emboj.7601566. Epub 2007 Feb 8. EMBO J. 2007. PMID: 17290221 Free PMC article. - Comparative genomics of cyclin-dependent kinases suggest co-evolution of the RNAP II C-terminal domain and CTD-directed CDKs.
Guo Z, Stiller JW. Guo Z, et al. BMC Genomics. 2004 Sep 20;5:69. doi: 10.1186/1471-2164-5-69. BMC Genomics. 2004. PMID: 15380029 Free PMC article. - The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress.
Mathur S, Vyas S, Kapoor S, Tyagi AK. Mathur S, et al. Plant Physiol. 2011 Dec;157(4):1609-27. doi: 10.1104/pp.111.188300. Epub 2011 Oct 21. Plant Physiol. 2011. PMID: 22021418 Free PMC article. - Mediator kinase module and human tumorigenesis.
Clark AD, Oldenbroek M, Boyer TG. Clark AD, et al. Crit Rev Biochem Mol Biol. 2015;50(5):393-426. doi: 10.3109/10409238.2015.1064854. Epub 2015 Jul 16. Crit Rev Biochem Mol Biol. 2015. PMID: 26182352 Free PMC article. Review. - Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man.
Boube M, Joulia L, Cribbs DL, Bourbon HM. Boube M, et al. Cell. 2002 Jul 26;110(2):143-51. doi: 10.1016/s0092-8674(02)00830-9. Cell. 2002. PMID: 12150923 Review.
Cited by
- All-Atomic Molecular Dynamic Studies of Human and Drosophila CDK8: Insights into Their Kinase Domains, the LXXLL Motifs, and Drug Binding Site.
Xu W, Xie XJ, Faust AK, Liu M, Li X, Chen F, Naquin AA, Walton AC, Kishbaugh PW, Ji JY. Xu W, et al. Int J Mol Sci. 2020 Oct 12;21(20):7511. doi: 10.3390/ijms21207511. Int J Mol Sci. 2020. PMID: 33053834 Free PMC article. - Differential Requirements for Mediator Complex Subunits in Drosophila melanogaster Host Defense Against Fungal and Bacterial Pathogens.
Huang C, Xu R, Liégeois S, Chen D, Li Z, Ferrandon D. Huang C, et al. Front Immunol. 2021 Mar 5;11:478958. doi: 10.3389/fimmu.2020.478958. eCollection 2020. Front Immunol. 2021. PMID: 33746938 Free PMC article. - Proteasome-mediated turnover of Arabidopsis MED25 is coupled to the activation of FLOWERING LOCUS T transcription.
Iñigo S, Giraldez AN, Chory J, Cerdán PD. Iñigo S, et al. Plant Physiol. 2012 Nov;160(3):1662-73. doi: 10.1104/pp.112.205500. Epub 2012 Sep 19. Plant Physiol. 2012. PMID: 22992513 Free PMC article. - Mediator Subunits MED16, MED14, and MED2 Are Required for Activation of ABRE-Dependent Transcription in Arabidopsis.
Lee M, Dominguez-Ferreras A, Kaliyadasa E, Huang WJ, Antony E, Stevenson T, Lehmann S, Schäfer P, Knight MR, Ntoukakis V, Knight H. Lee M, et al. Front Plant Sci. 2021 Mar 11;12:649720. doi: 10.3389/fpls.2021.649720. eCollection 2021. Front Plant Sci. 2021. PMID: 33777083 Free PMC article. - Synergistic repression of thyroid hyperplasia by cyclin C and Pten.
Jezek J, Wang K, Yan R, Di Cristofano A, Cooper KF, Strich R. Jezek J, et al. J Cell Sci. 2019 Aug 15;132(16):jcs230029. doi: 10.1242/jcs.230029. J Cell Sci. 2019. PMID: 31331961 Free PMC article.
References
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. - PubMed
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. - PubMed
- Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. - PubMed
- Kornberg RD. The eukaryotic gene transcription machinery. Biol. Chem. 2001;382:1103–1107. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous