Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications - PubMed (original) (raw)
. 2008 Jun 1;14(11):3380-5.
doi: 10.1158/1078-0432.CCR-07-5194.
Sven Perner, Elizabeth M Genega, Martin Sanda, Matthias D Hofer, Kirsten D Mertz, Pamela L Paris, Jeff Simko, Tarek A Bismar, Gustavo Ayala, Rajal B Shah, Massimo Loda, Mark A Rubin
Affiliations
- PMID: 18519767
- PMCID: PMC3717517
- DOI: 10.1158/1078-0432.CCR-07-5194
Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications
Juan-Miguel Mosquera et al. Clin Cancer Res. 2008.
Abstract
Purpose: More than 1,300,000 prostate needle biopsies are done annually in the United States with up to 16% incidence of isolated high-grade prostatic intraepithelial neoplasia (HGPIN). HGPIN has low predictive value for identifying prostate cancer on subsequent needle biopsies in prostate-specific antigen-screened populations. In contemporary series, prostate cancer is detected in approximately 20% of repeat biopsies following a diagnosis of HGPIN. Further, discrete histologic subtypes of HGPIN with clinical implication in management have not been characterized. The TMPRSS2-ERG gene fusion that has recently been described in prostate cancer has also been shown to occur in a subset of HGPIN. This may have significant clinical implications given that TMPRSS2-ERG fusion prostate cancer is associated with a more aggressive clinical course.
Experimental design: In this study, we assessed a series of HGPIN lesions and paired prostate cancer for the presence of TMPRSS2-ERG gene fusion.
Results: Fusion-positive HGPIN was observed in 16% of the 143 number of lesions, and in all instances, the matching cancer shared the same fusion pattern. Sixty percent of TMPRSS2-ERG fusion prostate cancer had fusion-negative HGPIN.
Conclusions: Given the more aggressive nature of TMPRSS2-ERG prostate cancer, the findings of this study raise the possibility that gene fusion-positive HGPIN lesions are harbingers of more aggressive disease. To date, pathologic, molecular, and clinical variables do not help stratify which men with HGPIN are at increased risk for a cancer diagnosis. Our results suggest that the detection of isolated TMPRSS2-ERG fusion HGPIN would improve the positive predictive value of finding TMPRSS2-ERG fusion prostate cancer in subsequent biopsies.
Figures
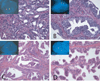
Figure 1. H&E stains and corresponding FISH images of TMPRSS2-ERG fusion assay
A: TMPRSS2-ERG fusion prostate cancer, Gleason grade 3+4=7. The inset picture shows a nucleus with one yellow and one red signal, demonstrating the presence of TMPRSS2-ERG fusion through deletion. B: Paired HGPIN lesion of prostate cancer in A. The HGPIN features tufting morphology. The inset picture shows a nucleus with two yellow signals, demonstrating absence of genetic aberration. C: TMPRSS2-ERG fusion prostate cancer, Gleason grade 4+4=8. The prostatectomy on this case showed predominant cribriform morphology. The inset picture shows a nucleus with one yellow and one red signal, demonstrating the presence of TMPRSS2-ERG fusion through deletion. D: Paired HGPIN lesion of prostate cancer in C. The HGPIN features tufting and micropapillary morphology. The inset picture shows a nucleus with the same pattern as the matching prostate cancer, demonstrating the presence of TMPRSS2-ERG fusion. Original magnification of H&E images, 20× objective. Original magnification of FISH images, 60× objective.
Figure 2. H&E stain and corresponding FISH image of TMPRSS2-ERG fusion assay
A: HGPIN lesion with adjacent atypical small acinar proliferation. This may represent either outpouching area or tangential section of HGPIN, or true early invasive adenocarcinoma. The red arrow points this area. The inset picture shows a nucleus with one yellow and one red signal, demonstrating the presence of TMPRSS2-ERG fusion through deletion. Original magnification of H&E images, 20× objective. Original magnification of FISH images, 60× objective. B: HGPIN and normal prostatic epithelium in the same gland. Red and green arrows point representative areas of HGPIN and normal prostatic epithelium, respectively. The inset pictures show a nucleus of normal epithelium with juxtaposed red-green signal pair (upper left), and a nucleus of HGPIN with one yellow and one red signal, demonstrating TMPRSS2-ERG fusion through deletion (lower right). The surrounding prostatic cancer, mostly Gleason pattern 4, also shared the same gene fusion pattern. Original magnification of H&E images, 20× objective. Original magnification of FISH images, 60× objective.
Similar articles
- TMPRSS2:ERG gene fusion predicts subsequent detection of prostate cancer in patients with high-grade prostatic intraepithelial neoplasia.
Park K, Dalton JT, Narayanan R, Barbieri CE, Hancock ML, Bostwick DG, Steiner MS, Rubin MA. Park K, et al. J Clin Oncol. 2014 Jan 20;32(3):206-11. doi: 10.1200/JCO.2013.49.8386. Epub 2013 Dec 2. J Clin Oncol. 2014. PMID: 24297949 Free PMC article. Clinical Trial. - Molecular evidence that invasive adenocarcinoma can mimic prostatic intraepithelial neoplasia (PIN) and intraductal carcinoma through retrograde glandular colonization.
Haffner MC, Weier C, Xu MM, Vaghasia A, Gürel B, Gümüşkaya B, Esopi DM, Fedor H, Tan HL, Kulac I, Hicks J, Isaacs WB, Lotan TL, Nelson WG, Yegnasubramanian S, De Marzo AM. Haffner MC, et al. J Pathol. 2016 Jan;238(1):31-41. doi: 10.1002/path.4628. Epub 2015 Oct 14. J Pathol. 2016. PMID: 26331372 Free PMC article. - Expression of ERG protein, a prostate cancer specific marker, in high grade prostatic intraepithelial neoplasia (HGPIN): lack of utility to stratify cancer risks associated with HGPIN.
He H, Osunkoya AO, Carver P, Falzarano S, Klein E, Magi-Galluzzi C, Zhou M. He H, et al. BJU Int. 2012 Dec;110(11 Pt B):E751-5. doi: 10.1111/j.1464-410X.2012.11557.x. Epub 2012 Oct 9. BJU Int. 2012. PMID: 23046279 - Prostate needle biopsies containing prostatic intraepithelial neoplasia or atypical foci suspicious for carcinoma: implications for patient care.
Epstein JI, Herawi M. Epstein JI, et al. J Urol. 2006 Mar;175(3 Pt 1):820-34. doi: 10.1016/S0022-5347(05)00337-X. J Urol. 2006. PMID: 16469560 Review. - Clinical applications of novel ERG immunohistochemistry in prostate cancer diagnosis and management.
Shah RB. Shah RB. Adv Anat Pathol. 2013 Mar;20(2):117-24. doi: 10.1097/PAP.0b013e3182862ac5. Adv Anat Pathol. 2013. PMID: 23399797 Review.
Cited by
- ETS-related gene (ERG) undermines genome stability in mouse prostate progenitors via Gsk3β dependent Nkx3.1 degradation.
Lorenzoni M, De Felice D, Beccaceci G, Di Donato G, Foletto V, Genovesi S, Bertossi A, Cambuli F, Lorenzin F, Savino A, Avalle L, Cimadamore A, Montironi R, Weber V, Carbone FG, Barbareschi M, Demichelis F, Romanel A, Poli V, Del Sal G, Julio MK, Gaspari M, Alaimo A, Lunardi A. Lorenzoni M, et al. Cancer Lett. 2022 May 28;534:215612. doi: 10.1016/j.canlet.2022.215612. Epub 2022 Mar 5. Cancer Lett. 2022. PMID: 35259458 Free PMC article. - ERG-TMPRSS2 rearrangement is shared by concurrent prostatic adenocarcinoma and prostatic small cell carcinoma and absent in small cell carcinoma of the urinary bladder: evidence supporting monoclonal origin.
Williamson SR, Zhang S, Yao JL, Huang J, Lopez-Beltran A, Shen S, Osunkoya AO, MacLennan GT, Montironi R, Cheng L. Williamson SR, et al. Mod Pathol. 2011 Aug;24(8):1120-7. doi: 10.1038/modpathol.2011.56. Epub 2011 Apr 15. Mod Pathol. 2011. PMID: 21499238 Free PMC article. - Anti-inflammatory drugs, antioxidants, and prostate cancer prevention.
Bardia A, Platz EA, Yegnasubramanian S, De Marzo AM, Nelson WG. Bardia A, et al. Curr Opin Pharmacol. 2009 Aug;9(4):419-26. doi: 10.1016/j.coph.2009.06.002. Epub 2009 Jul 1. Curr Opin Pharmacol. 2009. PMID: 19574101 Free PMC article. Review. - [High-grade prostatic intraepithelial neoplasia: the only accepted prostate cancer precursor lesion].
Braun M, Perner S. Braun M, et al. Pathologe. 2011 Nov;32 Suppl 2:237-41. doi: 10.1007/s00292-011-1500-9. Pathologe. 2011. PMID: 21845356 Review. German. - Correlation of urine TMPRSS2:ERG and PCA3 to ERG+ and total prostate cancer burden.
Young A, Palanisamy N, Siddiqui J, Wood DP, Wei JT, Chinnaiyan AM, Kunju LP, Tomlins SA. Young A, et al. Am J Clin Pathol. 2012 Nov;138(5):685-96. doi: 10.1309/AJCPU7PPWUPYG8OH. Am J Clin Pathol. 2012. PMID: 23086769 Free PMC article.
References
- Bostwick DG, Qian J, Frankel K. The incidence of high grade prostatic intraepithelial neoplasia in needle biopsies. J Urol. 1995;154(5):1791–1794. - PubMed
- Langer JE, Rovner ES, Coleman BG, et al. Strategy for repeat biopsy of patients with prostatic intraepithelial neoplasia detected by prostate needle biopsy. J Urol. 1996;155(1):228–231. - PubMed
- Moore CK, Karikehalli S, Nazeer T, Fisher HA, Kaufman RP, Jr, Mian BM. Prognostic significance of high grade prostatic intraepithelial neoplasia and atypical small acinar proliferation in the contemporary era. J Urol. 2005;173(1):70–72. - PubMed
- Naya Y, Ayala AG, Tamboli P, Babaian RJ. Can the number of cores with high-grade prostate intraepithelial neoplasia predict cancer in men who undergo repeat biopsy? Urology. 2004;63(3):503–508. - PubMed
- Vis AN, Van Der Kwast TH. Prostatic intraepithelial neoplasia and putative precursor lesions of prostate cancer: a clinical perspective. BJU Int. 2001;88(2):147–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01AG21404/AG/NIA NIH HHS/United States
- P50 CA090381/CA/NCI NIH HHS/United States
- R01 AG021404/AG/NIA NIH HHS/United States
- UO1 CA113913/CA/NCI NIH HHS/United States
- U01 CA113913/CA/NCI NIH HHS/United States
- P50 CA089520/CA/NCI NIH HHS/United States
- P50CA89520/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical