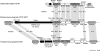Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis - PubMed (original) (raw)
Comparative Study
doi: 10.1093/dnares/dsn013. Epub 2008 Jun 3.
Hideki Hirakawa, Atsushi Yamashita, Naoya Ohara, Mikio Shoji, Hideharu Yukitake, Keisuke Nakayama, Hidehiro Toh, Fuminobu Yoshimura, Satoru Kuhara, Masahira Hattori, Tetsuya Hayashi, Koji Nakayama
Affiliations
- PMID: 18524787
- PMCID: PMC2575886
- DOI: 10.1093/dnares/dsn013
Comparative Study
Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis
Mariko Naito et al. DNA Res. 2008 Aug.
Abstract
The gram-negative anaerobic bacterium Porphyromonas gingivalis is a major causative agent of chronic periodontitis. Porphyromonas gingivalis strains have been classified into virulent and less-virulent strains by mouse subcutaneous soft tissue abscess model analysis. Here, we present the whole genome sequence of P. gingivalis ATCC 33277, which is classified as a less-virulent strain. We identified 2090 protein-coding sequences (CDSs), 4 RNA operons, and 53 tRNA genes in the ATCC 33277 genome. By genomic comparison with the virulent strain W83, we identified 461 ATCC 33277-specific and 415 W83-specific CDSs. Extensive genomic rearrangements were observed between the two strains: 175 regions in which genomic rearrangements have occurred were identified. Thirty-five of those genomic rearrangements were inversion or translocation and 140 were simple insertion, deletion, or replacement. Both strains contained large numbers of mobile elements, such as insertion sequences, miniature inverted-repeat transposable elements (MITEs), and conjugative transposons, which are frequently associated with genomic rearrangements. These findings indicate that the mobile genetic elements have been deeply involved in the extensive genome rearrangement of P. gingivalis and the occurrence of many of the strain-specific CDSs. We also describe here a very unique feature of MITE400, which we renamed MITEPgRS (MITE of P. gingivalis with Repeating Sequences).
Figures
Figure 1
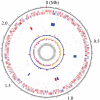
Circular map of the chromosome of P. gingivalis strain ATCC 33277. From the outside, the first and second circles show CDSs on the plus and minus strands, respectively. CDSs conserved in strains ATCC 33277 and W83 are indicated in red and ATCC 33277-specific CDSs in blue. The 3rd to 5th circles show IS elements (orange, IS_Pg1_; light green, IS_Pg2_; magenta, IS_Pg3_; cyan, IS_Pg4_; brown, IS_Pg5_; blue, IS_Pg6_), MITEs (magenta, MITE239; black, MITE_PgRS_; cyan, MITE700), CTns, and Tns (blue, CTnPg1-a, CTnPg1-b, CTnPg2, and CTnPg3; red, Tn_Pg17_), respectively. The 6th and 7th circles show rrn operons and tRNA genes, respectively. The 8th circle shows the result of _χ_2 analysis of nucleotide composition. Regions exhibiting values of >600 are indicated in red and those of <600 are indicated in blue. The G + C skew and G + C content are shown in the 9th and 10th circles, respectively.
Figure 2
Novel CTns and Tn identified in the ATCC 33277 genome. (A) Structures of CTnPg1-a, CTnPg1-b, CTnPg2, and CTnPg3. (B) Structure of Tn_Pg17_. CDSs are depicted by arrows and IS elements by open boxes (vertically striped arrow, tra or mob genes; thin arrow in box, IS transposase; black arrow, partial transposase; hatched arrow, other functionally annotated CDS; white arrow, hypothetical protein). Black triangles in CTnPg1-a and CTnPg1-b indicate direct repeat sequences, and black boxes in Tn_Pg17_ MITEs. The regions of CTnPg1-a and CTnPg1-b indicated by gray shading have an identical sequence.
Figure 3
MITE in P. gingivalis with Repeating Structure (MITE_PgRS_). (A) Schematic presentation of the consensus structure of MITE_PgRS_ and the structures of 20 copies of MITE_PgRS_ identified in the ATCC 33277 genome are shown. Three kind of repeat sequences, Repeats A, B, and C, are depicted by colored boxes. Red triangles indicate IR sequences and a black thick line in MITE_PgRS__08 a unique nucleotide sequence. (B) Consensus sequences of Repeats A, B, and C are shown.
Figure 4
The DNA sequence identity plot of P. gingivalis ATCC 33277 and W83 chromosomes. The dnaA gene is located at the left and bottom corner. Black circles indicate mobile genetic elements (CTn Tn, IS, MITE, or a not-well-defined large mobile element of W83). The chromosomal locations of other genetic elements that mediated inversions or translocations are shown in the right: rrn operons (black squares), duplicated regions coding for a histone-like DNA binding protein, a hypothetical protein and elongation factor P (open squares), 12/13 bp repeat sequences (black triangle), and 11 bp repeat sequences (open triangle).
Figure 5
Comparison of CRISPR-30-36 regions of P. gingivalis and B. fragilis. Locations and directions of CDSs (arrows) and repeat regions (black rectangles) are drawn to scale. Homologous CDSs are indicated by gray shading, and their amino-acid sequence identities are also shown. CDSs for IS transposases are indicated by black arrows, cas genes by vertically striped arrows, other functionally annotated CDSs by hatched arrows, and CDSs for hypothetical proteins by white arrows. The identity between PGN_1964 and PG2016 is 15.7%.
Figure 6
Excision of CTnPg1-a. (A) Schematic presentation of the structure of CTnPg1-a and the strategy to detect the excised circular intermediate and cast-off chromosome. Locations of PCR primers are indicated by black arrow heads. CDSs on CTnPg1 are depicted by black arrows and other CDSs by open arrows. Open triangles indicate att regions of CTnPg1. (B) Agarose gel electrophoresis of PCR products obtained by the primer pairs CTnPg1-right/CTnPg1-left (lane 1) and CTnPg1-up/CTnPg1-down (lane 2). (C) Sequence alignment of _att_P, attB, attL, and attR regions of CTnPg1. The 14 bp core sequence is indicated by a box.
Similar articles
- Characterization of the Porphyromonas gingivalis conjugative transposon CTnPg1: determination of the integration site and the genes essential for conjugal transfer.
Naito M, Sato K, Shoji M, Yukitake H, Ogura Y, Hayashi T, Nakayama K. Naito M, et al. Microbiology (Reading). 2011 Jul;157(Pt 7):2022-2032. doi: 10.1099/mic.0.047803-0. Epub 2011 Apr 28. Microbiology (Reading). 2011. PMID: 21527470 - Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis.
Chen T, Hosogi Y, Nishikawa K, Abbey K, Fleischmann RD, Walling J, Duncan MJ. Chen T, et al. J Bacteriol. 2004 Aug;186(16):5473-9. doi: 10.1128/JB.186.16.5473-5479.2004. J Bacteriol. 2004. PMID: 15292149 Free PMC article. - The core genome of the anaerobic oral pathogenic bacterium Porphyromonas gingivalis.
Brunner J, Wittink FR, Jonker MJ, de Jong M, Breit TM, Laine ML, de Soet JJ, Crielaard W. Brunner J, et al. BMC Microbiol. 2010 Sep 29;10:252. doi: 10.1186/1471-2180-10-252. BMC Microbiol. 2010. PMID: 20920246 Free PMC article. - Identification by subtractive hybridization of a novel insertion sequence specific for virulent strains of Porphyromonas gingivalis.
Sawada K, Kokeguchi S, Hongyo H, Sawada S, Miyamoto M, Maeda H, Nishimura F, Takashiba S, Murayama Y. Sawada K, et al. Infect Immun. 1999 Nov;67(11):5621-5. doi: 10.1128/IAI.67.11.5621-5625.1999. Infect Immun. 1999. PMID: 10531208 Free PMC article. - Iron and heme utilization in Porphyromonas gingivalis.
Olczak T, Simpson W, Liu X, Genco CA. Olczak T, et al. FEMS Microbiol Rev. 2005 Jan;29(1):119-44. doi: 10.1016/j.femsre.2004.09.001. FEMS Microbiol Rev. 2005. PMID: 15652979 Review.
Cited by
- In Search of Spectroscopic Signatures of Periodontitis: A SERS-Based Magnetomicrofluidic Sensor for Detection of Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans.
Witkowska E, Łasica AM, Niciński K, Potempa J, Kamińska A. Witkowska E, et al. ACS Sens. 2021 Apr 23;6(4):1621-1635. doi: 10.1021/acssensors.1c00166. Epub 2021 Apr 1. ACS Sens. 2021. PMID: 33792284 Free PMC article. - Dipeptidyl-peptidases: Key enzymes producing entry forms of extracellular proteins in asaccharolytic periodontopathic bacterium Porphyromonas gingivalis.
Nemoto TK, Ohara Nemoto Y. Nemoto TK, et al. Mol Oral Microbiol. 2021 Apr;36(2):145-156. doi: 10.1111/omi.12317. Epub 2020 Oct 12. Mol Oral Microbiol. 2021. PMID: 33006264 Free PMC article. Review. - Lysine gingipain (kgp) biovars of Porphyromonas gingivalis exhibit differential distribution on oral mucosal sites.
Nadkarni MA, Chhour KL, Browne G, Jacques NA, Hunter N. Nadkarni MA, et al. J Clin Microbiol. 2009 Oct;47(10):3350-2. doi: 10.1128/JCM.00753-09. Epub 2009 Aug 12. J Clin Microbiol. 2009. PMID: 19675219 Free PMC article. - Oral microbiome diversity in chimpanzees from Gombe National Park.
Ozga AT, Gilby I, Nockerts RS, Wilson ML, Pusey A, Stone AC. Ozga AT, et al. Sci Rep. 2019 Nov 22;9(1):17354. doi: 10.1038/s41598-019-53802-1. Sci Rep. 2019. PMID: 31758037 Free PMC article. - Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics?
Hajishengallis G. Hajishengallis G. Microbes Infect. 2009 May-Jun;11(6-7):637-45. doi: 10.1016/j.micinf.2009.03.009. Epub 2009 Apr 5. Microbes Infect. 2009. PMID: 19348960 Free PMC article. Review.
References
- Papapanou P. N. Epidemiology of periodontal diseases: an update. J. Int. Acad. Periodontol. 1999;1:110–116. - PubMed
- Irfan U. M., Dawson D. V., Bissada N. F. Epidemiology of periodontal disease: a review and clinical perspectives. J. Int. Acad. Periodontol. 2001;3:14–21. - PubMed
- Armitage G. C. Periodontal diseases: diagnosis. Ann. Periodontol. 1996;1:37–215. - PubMed
- Oliver R. C., Brown L. J., Loe H. Periodontal diseases in the United States population. J. Periodontol. 1998;69:269–278. - PubMed
- Mattila K. J., Valtonen V. V., Nieminen M., Huttunen J. K. Dental infection and the risk of new coronary events: prospective study of patients with documented coronary artery disease. Clin. Infect. Dis. 1995;20:588–592. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases