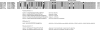Sensitive detection of multiple rotavirus genotypes with a single reverse transcription-real-time quantitative PCR assay - PubMed (original) (raw)
Sensitive detection of multiple rotavirus genotypes with a single reverse transcription-real-time quantitative PCR assay
Ion Gutiérrez-Aguirre et al. J Clin Microbiol. 2008 Aug.
Abstract
Rotaviruses are one of the major causes of diarrhea in infants and children under 5 years old, especially affecting developing countries. In natural disasters, fecal matter and potable waters can mix, allowing low, yet infective, concentrations of rotavirus to be present in water supplies, constituting a risk for the population. Any of the most commonly detected rotavirus genotypes could originate an outbreak. The development of a fast and sensitive method that could detect the broadest possible range of rotavirus genotypes would help with efficient diagnosis and prevention. We have designed a reverse transcription (RT)-real-time quantitative PCR approach targeted to the rotaviral VP2 gene, based on a multiple-sequence alignment of different human rotaviral strains. To overcome the high nucleotide sequence diversity, multiple forward and reverse primers were used, in addition to a degenerate probe. The performance of the assay was tested on isolates representing the most prevalent human genotypes: G1P[8], G2P[4], G3P[8], G4P[8], G9P[8], and G12P[8]. The developed method improved classical rotavirus detection by enzyme-linked immunosorbent assay and nested RT-PCR by 5 and at least 1 order of magnitude, respectively. A survey of 159 stool samples indicated that the method can efficiently detect a broad range of rotavirus strains, including different G-P genotype combinations of human, porcine, and bovine origin. No cross-reactivity was observed with other enteric viruses, such as astrovirus, sapovirus, and norovirus.
Figures
FIG. 1.
Design of the qPCR approach. The sequences of the five forward primers, two reverse primers, and degenerate TaqMan MGB probe are shown, followed by the sequence accession numbers and positions to which each primer would anneal without any nucleotide mismatch. The corresponding accession numbers and rotavirus genotype for each of the aligned sequences are shown to the left of the alignment.
Similar articles
- Detection and genotyping of human rotavirus VP4 and VP7 genes by reverse transcriptase PCR and reverse hybridization.
van Doorn LJ, Kleter B, Hoefnagel E, Stainier I, Poliszczak A, Colau B, Quint W. van Doorn LJ, et al. J Clin Microbiol. 2009 Sep;47(9):2704-12. doi: 10.1128/JCM.00378-09. Epub 2009 Jun 24. J Clin Microbiol. 2009. PMID: 19553575 Free PMC article. - Evaluation of a multiplex real time reverse transcription PCR assay for the detection and quantitation of the most common human rotavirus genotypes.
Kottaridi C, Spathis AT, Ntova CK, Papaevangelou V, Karakitsos P. Kottaridi C, et al. J Virol Methods. 2012 Mar;180(1-2):49-53. doi: 10.1016/j.jviromet.2011.12.009. Epub 2012 Jan 11. J Virol Methods. 2012. PMID: 22245180 - Increased sensitivity for various rotavirus genotypes in stool specimens by amending three mismatched nucleotides in the forward primer of a real-time RT-PCR assay.
Pang X, Cao M, Zhang M, Lee B. Pang X, et al. J Virol Methods. 2011 Mar;172(1-2):85-7. doi: 10.1016/j.jviromet.2010.12.013. Epub 2010 Dec 23. J Virol Methods. 2011. PMID: 21185331 - Detection and characterization of group A rotaviruses in children hospitalized with acute gastroenteritis in Norway, 2006-2008.
Vainio K, Nordbø SA, Njølstad G, Størvold G, Døllner H, Midgaard C, Bosse FJ, Rognlien AG, Rojahn A, Wathne KO, Flem E. Vainio K, et al. J Med Virol. 2009 Oct;81(10):1839-44. doi: 10.1002/jmv.21576. J Med Virol. 2009. PMID: 19697411 - Equine rotaviruses--current understanding and continuing challenges.
Bailey KE, Gilkerson JR, Browning GF. Bailey KE, et al. Vet Microbiol. 2013 Nov 29;167(1-2):135-44. doi: 10.1016/j.vetmic.2013.07.010. Epub 2013 Jul 22. Vet Microbiol. 2013. PMID: 23932076 Free PMC article. Review.
Cited by
- Prevalence and Genetic Diversity of Enteric Viruses in Children with Diarrhea in Ouagadougou, Burkina Faso.
Ouédraogo N, Kaplon J, Bonkoungou IJ, Traoré AS, Pothier P, Barro N, Ambert-Balay K. Ouédraogo N, et al. PLoS One. 2016 Apr 19;11(4):e0153652. doi: 10.1371/journal.pone.0153652. eCollection 2016. PLoS One. 2016. PMID: 27092779 Free PMC article. - Genomic revelations: investigating rotavirus a presence in wild ruminants and its zoonotic potential.
Šenica P, Žele Vengušt D, Vengušt G, Kuhar U. Šenica P, et al. Front Vet Sci. 2024 Aug 15;11:1429654. doi: 10.3389/fvets.2024.1429654. eCollection 2024. Front Vet Sci. 2024. PMID: 39211480 Free PMC article. - Prevalence of major enteric pathogens in Australian dairy calves with diarrhoea.
Izzo MM, Kirkland PD, Mohler VL, Perkins NR, Gunn AA, House JK. Izzo MM, et al. Aust Vet J. 2011 May;89(5):167-73. doi: 10.1111/j.1751-0813.2011.00692.x. Aust Vet J. 2011. PMID: 21495987 Free PMC article. - A Remarkable Genetic Diversity of Rotavirus A Circulating in Red Fox Population in Croatia.
Čolić D, Krešić N, Mihaljević Ž, Andreanszky T, Balić D, Lolić M, Brnić D. Čolić D, et al. Pathogens. 2021 Apr 16;10(4):485. doi: 10.3390/pathogens10040485. Pathogens. 2021. PMID: 33923799 Free PMC article. - One-step RT-droplet digital PCR: a breakthrough in the quantification of waterborne RNA viruses.
Rački N, Morisset D, Gutierrez-Aguirre I, Ravnikar M. Rački N, et al. Anal Bioanal Chem. 2014 Jan;406(3):661-7. doi: 10.1007/s00216-013-7476-y. Epub 2013 Nov 26. Anal Bioanal Chem. 2014. PMID: 24276251 Free PMC article.
References
- Amar, C. F. L., C. L. East, J. Gray, M. Iturriza-Gómara, E. A. Maclure, and J. McLauchlin. 2007. Detection by PCR of eight groups of enteric pathogens in 4,627 faecal samples: re-examination of the English case-control Infectious Intestinal Disease Study (1993-1996). Eur. J. Microbiol. Infect. Dis. 26311-323. - PubMed
- Boben, J., P. Kramberger, N. Petrovič, K. Cankar, M. Peterka, A. Štrancar, and M. Ravnikar. 2007. Detection and quantification of tomato mosaic virus in irrigation waters. Eur. J. Plant Pathol. 11859-71.
- Burns, M. J., H. Valdivia, and N. Harris. 2004. Analysis and interpretation of data from real-time PCR trace detection methods using quantitation of GM soya as a model system. Anal. Bioanal. Chem. 3781616-1623. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical