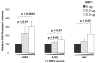LINE-1 ORF1 protein enhances Alu SINE retrotransposition - PubMed (original) (raw)
LINE-1 ORF1 protein enhances Alu SINE retrotransposition
Nicholas Wallace et al. Gene. 2008.
Abstract
Retroelements have contributed over one third of the human genome mass. The currently active LINE-1 (L1) codes for two proteins (ORF1p and ORF2p), both strictly required for retrotransposition. In contrast, the non-coding parasitic SINE (Alu) only appears to need the L1 ORF2p for its own amplification. This requirement was previously determined using a tissue culture assay system in human cells (HeLa). Because HeLa are likely to express functional L1 proteins, it is possible that low levels of endogenous ORF1p are necessary for the observed tagged Alu mobilization. By individually expressing ORF1 and ORF2 proteins from both human (L1RP and LRE3) and rodent (L1A102 and L1spa) L1 sources, we demonstrate that increasing amounts of ORF1 expressing vector enhances tagged Alu mobilization in HeLa cells. In addition, using chicken fibroblast cells as an alternate cell culture source, we confirmed that ORF1p is not strictly required for Alu mobilization in our assay. Supporting our observations in HeLa cells, we find that tagged Alu retrotransposition is improved by supplementation of ORF1p in the cultured chicken cells. We postulate that L1 ORF1p plays either a direct or indirect role in enhancing the interaction between the Alu RNA and the required factors needed for its retrotransposition.
Figures
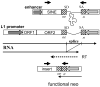
Figure 1. Schematic of the LINE and SINE assay
RNA transcription is performed by L1 5′UTR promoter or the internal pol III promoter of the SINE enhanced by the 7SL upstream sequence. An intron (a self-splicing in the SINE vector) interrupts the neomycin (neo) resistance gene (hatched box) and promoter present in an inverted orientation. Because of orientation, the intron will splice out only from the transcripts generated by the retroelement’s promoter. The RNA is reverse transcribed, followed by integration of the cDNA into the genome. The new insert contains a functional neomycin gene.
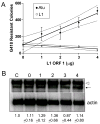
Figure 2. L1 ORF1p enhances retrotransposition of a tagged Alu but not L1
A. L1 ORF1p gradient and retrotransposition activity of L1 and Alu (driven by ORF2RP) in HeLa cells. Cells were transfected with L1-tag (open circle) or Alu_neo_TET plus ORF2 (black circle) and supplemented with increasing concentrations of ORF1. The mean G418R colonies for each retroelement are plotted as a solid line, where error bars represent the standard deviation and the flanking discontinuous lines represent the 95% confidence interval. The calculated slope for Alu is 95.12 ± 11.92 and for L1 -12.70 ± 3.55 and shown to be significantly different as determined by Least squares regression analysis, p = 0.00013 n = 3. B. Alu-tag RNA transcription and processing is unaffected by the supplementation of ORF1p. Representative northern blot analysis of polyA selected RNA from transfected cells with the tagged Alu vector alone (C, control), or supplemented with ORF2p plus different amounts of ORF1p (0 – 4 μg). The unspliced (open arrowhead), spliced RNA (small arrow) and actin bands are indicated. Four separate transfections and northern blot analyses were performed for the quantitative evaluation. The numbers below represent the relative mean ± SD of Alu-tag spliced (β-actin corrected) relative to the control (C) which was arbitrarily defined as “1”. No significant differences between the amounts of the spliced Alu-tag RNA of any of the experimental conditions relative to the control (Student paired t-test p ≥ 0.23).
Figure 3. Both human and rodent ORF1p enhance ORF2 driven Alu retrotransposition
HeLa cells were transfected with the tagged Alu supplemented with an ORF2p expression vector from the human L1LRE element or the rodent L1A102 or L1spa elements (n = 5). Each Alu-ORF2p set was cotransfected with different amounts of the corresponding human/rodent ORF1p expression vector or empty vector for the “0 μg” or ORF2 only control. The relative G418R colonies were graphed using the ORF2 only control (black column) reference, which was arbitrarily assigned as 100. The p-values (Student paired t-test) for significant differences are indicated above the columns.
Figure 4. A ORF2p is sufficient to mobilize SINEs in chicken cells
A. Retrotransposition activity of tagged Alu supplemented with the synthetic ORF2s from the human L1RP or the mouse L1spa in chicken embryo fibroblast cells. The mean of the total number of G418 resistant colonies for each time point is shown (n= 5). No colonies were ever observed for the Alu-tag cotransfected with the empty vector (control) indicated by “0”. B. Evaluation of the Alu inserts from the chicken cells. Alu inserts were evaluated by PCR analysis using primers to the Alu sequence and the neomycin gene sequence (shown as bold arrows in Figure 1). DNA was recovered from pooled G418 resistance colonies generated in the chicken embryo fibroblast cells transfected with the tagged Alu vector plus the synthetic versions of either human (L1RP) or mouse (L1spa) ORF2 with and without supplementation of the corresponding ORF1p. An open arrowhead indicates the PCR product corresponding to an insert containing the spliced version (open arrowhead) of the Alu expression vector. The Alu expression plasmid (P lane) was used as the control for unspliced products (small arrow). DNA from untransfected cells were used as the negative control (−). M is DNA marker. C. ORF1p enhances SINE retrotransposition in chicken cells. Cells were transfected with the tagged Alu plus the human ORF2p (L1RP) supplemented with different amounts of the ORF1p expression vector (0, 1, 2 or 6 μg). Columns represent the mean number of G418 resistant colonies for each time point (n = 5) and S.D. shown as error bars. Results significantly different from the no ORF1 reference transfection with p-values of p≤ 0.05 (Student paired t-test) are indicated by an asterisk (*).
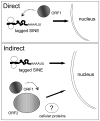
Figure 5. Model of potential roles of ORF1p in SINE retrotransposition
Although onlyORF2p is strictly required for SINE retrotransposition, supplementation with ORF1p enhances their mobilization. There are two potential scenarios of how ORF1p increases Alu retrotransposition. First, because of the chaperone nature of ORF1p, the direct interaction of ORF1p with the SINE RNA may play a role in protecting the transcript from degradation or aiding the RNP complex to reach the nucleus. Alternatively, in the second scenario, ORF1p may have an indirect role by interacting with other components such as ORF2p or cellular factors to facilitate retrotransposition. In addition, ORF1p could have a role in increasing the half life of ORF2p or targeting other cellular factors and ORF2p to the nucleus.
Similar articles
- The RNA polymerase dictates ORF1 requirement and timing of LINE and SINE retrotransposition.
Kroutter EN, Belancio VP, Wagstaff BJ, Roy-Engel AM. Kroutter EN, et al. PLoS Genet. 2009 Apr;5(4):e1000458. doi: 10.1371/journal.pgen.1000458. Epub 2009 Apr 24. PLoS Genet. 2009. PMID: 19390602 Free PMC article. - Molecular reconstruction of extinct LINE-1 elements and their interaction with nonautonomous elements.
Wagstaff BJ, Kroutter EN, Derbes RS, Belancio VP, Roy-Engel AM. Wagstaff BJ, et al. Mol Biol Evol. 2013 Jan;30(1):88-99. doi: 10.1093/molbev/mss202. Epub 2012 Aug 23. Mol Biol Evol. 2013. PMID: 22918960 Free PMC article. - Identification of L1 ORF2p sequence important to retrotransposition using Bipartile Alu retrotransposition (BAR).
Christian CM, deHaro D, Kines KJ, Sokolowski M, Belancio VP. Christian CM, et al. Nucleic Acids Res. 2016 Jun 2;44(10):4818-34. doi: 10.1093/nar/gkw277. Epub 2016 Apr 19. Nucleic Acids Res. 2016. PMID: 27095191 Free PMC article. - Protein-nucleic acid interactions of LINE-1 ORF1p.
Naufer MN, Furano AV, Williams MC. Naufer MN, et al. Semin Cell Dev Biol. 2019 Feb;86:140-149. doi: 10.1016/j.semcdb.2018.03.019. Epub 2018 Mar 31. Semin Cell Dev Biol. 2019. PMID: 29596909 Free PMC article. Review. - Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1.
Martin SL. Martin SL. RNA Biol. 2010 Nov-Dec;7(6):706-11. doi: 10.4161/rna.7.6.13766. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21045547 Free PMC article. Review.
Cited by
- LINEs, SINEs and other retroelements: do birds of a feather flock together?
Roy-Engel AM. Roy-Engel AM. Front Biosci (Landmark Ed). 2012 Jan 1;17(4):1345-61. doi: 10.2741/3991. Front Biosci (Landmark Ed). 2012. PMID: 22201808 Free PMC article. Review. - Induction of long interspersed nucleotide element-1 (L1) retrotransposition by 6-formylindolo[3,2-b]carbazole (FICZ), a tryptophan photoproduct.
Okudaira N, Iijima K, Koyama T, Minemoto Y, Kano S, Mimori A, Ishizaka Y. Okudaira N, et al. Proc Natl Acad Sci U S A. 2010 Oct 26;107(43):18487-92. doi: 10.1073/pnas.1001252107. Epub 2010 Sep 17. Proc Natl Acad Sci U S A. 2010. PMID: 20852066 Free PMC article. - ECAT11/L1td1 is enriched in ESCs and rapidly activated during iPSC generation, but it is dispensable for the maintenance and induction of pluripotency.
Iwabuchi KA, Yamakawa T, Sato Y, Ichisaka T, Takahashi K, Okita K, Yamanaka S. Iwabuchi KA, et al. PLoS One. 2011;6(5):e20461. doi: 10.1371/journal.pone.0020461. Epub 2011 May 26. PLoS One. 2011. PMID: 21637830 Free PMC article. - _ALU_minating the Path of Atherosclerosis Progression: Chaos Theory Suggests a Role for Alu Repeats in the Development of Atherosclerotic Vascular Disease.
Hueso M, Cruzado JM, Torras J, Navarro E. Hueso M, et al. Int J Mol Sci. 2018 Jun 12;19(6):1734. doi: 10.3390/ijms19061734. Int J Mol Sci. 2018. PMID: 29895733 Free PMC article. Review. - Dissolution of ribonucleoprotein condensates by the embryonic stem cell protein L1TD1.
Jin SW, Seong Y, Yoon D, Kwon YS, Song H. Jin SW, et al. Nucleic Acids Res. 2024 Apr 12;52(6):3310-3326. doi: 10.1093/nar/gkad1244. Nucleic Acids Res. 2024. PMID: 38165001 Free PMC article.
References
- Asch HL, Eliacin E, Fanning TG, Connolly JL, Bratthauer G, Asch BB. Comparative expression of the LINE-1 p40 protein in human breast carcinomas and normal breast tissues. Oncol Res. 1996;8:239–247. - PubMed
- Boissinot S, Furano AV. Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol. 2001;18:2186–2194. - PubMed
- Bratthauer GL, Fanning TG. Active LINE-1 retrotransposons in human testicular cancer. Oncogene. 1992;7:507–510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM079709/GM/NIGMS NIH HHS/United States
- P20 RR020152-01/RR/NCRR NIH HHS/United States
- R01 GM045668/GM/NIGMS NIH HHS/United States
- P20 RR020152/RR/NCRR NIH HHS/United States
- R01 GM079709-01A1/GM/NIGMS NIH HHS/United States
- R01GM45668/GM/NIGMS NIH HHS/United States
- P20RR020152/RR/NCRR NIH HHS/United States
- P20 RR020152-02/RR/NCRR NIH HHS/United States
- R01 GM045668-15/GM/NIGMS NIH HHS/United States
- R01GM79709/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources