Intersection of the RNA interference and X-inactivation pathways - PubMed (original) (raw)
Intersection of the RNA interference and X-inactivation pathways
Yuya Ogawa et al. Science. 2008.
Abstract
In mammals, dosage compensation is achieved by X-chromosome inactivation (XCI) in the female. The noncoding Xist gene initiates silencing of the X chromosome, whereas its antisense partner Tsix blocks silencing. The complementarity of Xist and Tsix RNAs has long suggested a role for RNA interference (RNAi). Here, we report that murine Xist and Tsix form duplexes in vivo. During XCI, the duplexes are processed to small RNAs (sRNAs), most likely on the active X (Xa) in a Dicer-dependent manner. Deleting Dicer compromises sRNA production and derepresses Xist. Furthermore, without Dicer, Xist RNA cannot accumulate and histone 3 lysine 27 trimethylation is blocked on the inactive X (Xi). The defects are partially rescued by truncating Tsix. Thus, XCI and RNAi intersect, down-regulating Xist on Xa and spreading silencing on Xi.
Figures
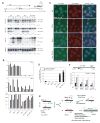
Figure 1. Small RNAs derived from Tsix/Xist
(A) xiRNAs from Xist repeat A (XA) region (map) detected by Northern analysis. Sense (s) and antisense (as) riboprobes detect Tsix and Xist, respectively. miR292-as controls are shown on same blots. M, male. F, female. (B) Northern analysis of xiRNAs from Xist exon 7. (C) Northern analysis of Xist promoter region. (D) Northern analysis of mutant cells. WT lanes identical to those in panel A (concurrent analysis).
Figure 2. Tsix and Xist RNA form long duplexes in vivo
(A) Map of Tsix/Xist and primer pairs (*). (B) In vivo RNAse protection assay. S, sense. AS, antisense. (C) Relative quantities of Xist and Tsix in duplexes measured at position 2 (bp 1206-1337 of Xist) by strand-specific real-time RT-PCR of protected RNA (RNAse+) as compared to total levels (RNAse–). Quantities are standardized to Xist in the Xist-Tsix duplex (Xist, RNAse+ = 1). Error bars=one standard deviation (SD) in triplicate reactions. (D) Quantities of protected Tsix or Xist RNAs (RNase+) relative to total Tsix or Xist (RNase-) for in vivo RNAse protection assays. Error bar=1SD, triplicate reactions. (E) In vivo RNAse protection assays to test allelic origin of dsRNA using strand-specific, allele-specific realtime RT-PCR with SNP-based primers for Xcas or Xmus alleles (Table). PCR of control genomic DNA shows high specificity (98% for mus, >99.99% for cas). Error bar=1SD, triplicate reactions. For test samples, the mus and cas fractions were amplified separately, normalized to genomic DNA (Xcas: Xmus =1:1), and plotted as a function of time. %cas = [Xcas RNA/ (Xcas RNA + Xmus RNA)] × 100. Because the bars show relative allelic fractions, quantities are only comparable within a single timepoint.
Figure 3. Dcr deficiency impairs xiRNA production and XCI
(A) Quantitative realtime RT-PCR of indicated transcripts normalized to β-actin. (B) Xist quantitation plotted on a log scale. (C,D) Northern analyses of miRNA292-as control (C) and xiRNAs (D) in mutant cells. Note accumulation of miRNA292-as precursors (*) in _Dcr_-/- cells. (E) Immuno-RNA FISH for Xist and H3-3meK27 on d10. DAPI, blue. (F) Phase contrast images of d10 EB.
Figure 4. Tsix genetically interacts with Dcr
(A) Allele-specific RT-PCR analysis. All RT- reactions were negative (not shown). Xist d0 samples (asterisks) were 10-fold overloaded to visualize low expression. (B) Realtime RT-PCR of indicated transcripts, each normalized to β-actin. (C) Immuno-RNA FISH for Xist and H3-3meK27 domains (arrowheads) on d10. (D) Frequency of aberrant H3-3meK27 enrichment in the Xist+ subpopulation of indicated cell lines. n=100-150. (E) Model: intersection of RNAi and XCI. (F) Methylation-sensitive restriction analysis of the Xist promoter. Genomic DNA was digested with EcoRV or EcoRV+HpaII. % uncut (methylated) DNA at HpaII is plotted.
Similar articles
- Synergy of Eed and Tsix in the repression of Xist gene and X-chromosome inactivation.
Shibata S, Yokota T, Wutz A. Shibata S, et al. EMBO J. 2008 Jul 9;27(13):1816-26. doi: 10.1038/emboj.2008.110. Epub 2008 May 29. EMBO J. 2008. PMID: 18511907 Free PMC article. - Uncoupling of X-linked gene silencing from XIST binding by DICER1 and chromatin modulation on human inactive X chromosome.
Kota SK, Roy Chowdhury D, Rao LK, Padmalatha V, Singh L, Bhadra U. Kota SK, et al. Chromosoma. 2015 Jun;124(2):249-62. doi: 10.1007/s00412-014-0495-4. Epub 2014 Nov 27. Chromosoma. 2015. PMID: 25428210 - X chromosome inactivation in the absence of Dicer.
Kanellopoulou C, Muljo SA, Dimitrov SD, Chen X, Colin C, Plath K, Livingston DM. Kanellopoulou C, et al. Proc Natl Acad Sci U S A. 2009 Jan 27;106(4):1122-7. doi: 10.1073/pnas.0812210106. Epub 2009 Jan 21. Proc Natl Acad Sci U S A. 2009. PMID: 19164542 Free PMC article. - X-chromosome inactivation: the molecular basis of silencing.
Panning B. Panning B. J Biol. 2008 Oct 27;7(8):30. doi: 10.1186/jbiol95. J Biol. 2008. PMID: 18983701 Free PMC article. Review. - X inactivation: Tsix and Xist as yin and yang.
Mlynarczyk SK, Panning B. Mlynarczyk SK, et al. Curr Biol. 2000 Dec 14-28;10(24):R899-903. doi: 10.1016/s0960-9822(00)00847-2. Curr Biol. 2000. PMID: 11137025 Review.
Cited by
- Balancing cohesin eviction and retention prevents aberrant chromosomal interactions, Polycomb-mediated repression, and X-inactivation.
Kriz AJ, Colognori D, Sunwoo H, Nabet B, Lee JT. Kriz AJ, et al. Mol Cell. 2021 May 6;81(9):1970-1987.e9. doi: 10.1016/j.molcel.2021.02.031. Epub 2021 Mar 15. Mol Cell. 2021. PMID: 33725485 Free PMC article. - Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis.
Fan J, Xing Y, Wen X, Jia R, Ni H, He J, Ding X, Pan H, Qian G, Ge S, Hoffman AR, Zhang H, Fan X. Fan J, et al. Genome Biol. 2015 Jul 14;16(1):139. doi: 10.1186/s13059-015-0705-2. Genome Biol. 2015. PMID: 26169368 Free PMC article. - Dynamic interplay and function of multiple noncoding genes governing X chromosome inactivation.
Yue M, Charles Richard JL, Ogawa Y. Yue M, et al. Biochim Biophys Acta. 2016 Jan;1859(1):112-20. doi: 10.1016/j.bbagrm.2015.07.015. Epub 2015 Aug 7. Biochim Biophys Acta. 2016. PMID: 26260844 Free PMC article. Review. - Random allelic expression in the adult human body.
Kravitz SN, Ferris E, Love MI, Thomas A, Quinlan AR, Gregg C. Kravitz SN, et al. Cell Rep. 2023 Jan 31;42(1):111945. doi: 10.1016/j.celrep.2022.111945. Epub 2023 Jan 5. Cell Rep. 2023. PMID: 36640362 Free PMC article. - Exploration of CTCF post-translation modifications uncovers Serine-224 phosphorylation by PLK1 at pericentric regions during the G2/M transition.
Del Rosario BC, Kriz AJ, Del Rosario AM, Anselmo A, Fry CJ, White FM, Sadreyev RI, Lee JT. Del Rosario BC, et al. Elife. 2019 Jan 24;8:e42341. doi: 10.7554/eLife.42341. Elife. 2019. PMID: 30676316 Free PMC article.
References
- Lyon MF. Nature. 1961;190:372. - PubMed
- Brown CJ, et al. Cell. 1992;71:527. - PubMed
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Nature. 1996;379:131. - PubMed
- Lee JT, Lu N. Cell. 1999;99:47. - PubMed
- Fire A, et al. Nature. 1998;391:806. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases