Patterns of evolution and host gene mimicry in influenza and other RNA viruses - PubMed (original) (raw)
Patterns of evolution and host gene mimicry in influenza and other RNA viruses
Benjamin D Greenbaum et al. PLoS Pathog. 2008.
Abstract
It is well known that the dinucleotide CpG is under-represented in the genomic DNA of many vertebrates. This is commonly thought to be due to the methylation of cytosine residues in this dinucleotide and the corresponding high rate of deamination of 5-methycytosine, which lowers the frequency of this dinucleotide in DNA. Surprisingly, many single-stranded RNA viruses that replicate in these vertebrate hosts also have a very low presence of CpG dinucleotides in their genomes. Viruses are obligate intracellular parasites and the evolution of a virus is inexorably linked to the nature and fate of its host. One therefore expects that virus and host genomes should have common features. In this work, we compare evolutionary patterns in the genomes of ssRNA viruses and their hosts. In particular, we have analyzed dinucleotide patterns and found that the same patterns are pervasively over- or under-represented in many RNA viruses and their hosts suggesting that many RNA viruses evolve by mimicking some of the features of their host's genes (DNA) and likely also their corresponding mRNAs. When a virus crosses a species barrier into a different host, the pressure to replicate, survive and adapt, leaves a footprint in dinucleotide frequencies. For instance, since human genes seem to be under higher pressure to eliminate CpG dinucleotide motifs than avian genes, this pressure might be reflected in the genomes of human viruses (DNA and RNA viruses) when compared to those of the same viruses replicating in avian hosts. To test this idea we have analyzed the evolution of the influenza virus since 1918. We find that the influenza A virus, which originated from an avian reservoir and has been replicating in humans over many generations, evolves in a direction strongly selected to reduce the frequency of CpG dinucleotides in its genome. Consistent with this observation, we find that the influenza B virus, which has spent much more time in the human population, has adapted to its human host and exhibits an extremely low CpG dinucleotide content. We believe that these observations directly show that the evolution of RNA viral genomes can be shaped by pressures observed in the host genome. As a possible explanation, we suggest that the strong selection pressures acting on these RNA viruses are most likely related to the innate immune response and to nucleotide motifs in the host DNA and RNAs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
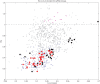
Figure 1. CpG odds ratio (η) versus C+G content for different ssRNA viruses.
A value of η close to 1 (marked in the figure with a dashed line), means that the measured value is very close to the expected, i.e. no significant pressure to create or eliminate CpGs. Most ssRNA viruses have strong pressure to eliminate CpGs. Human viruses, marked with black (ssRNA+) and red (ssRNA−) filled circles, are at the lower part of the CpG distribution for a given nucleotide content. In particular, the lowest ssRNA viruses in this set are Hepatitis A and Hemagglutinin in influenza C, both human viruses. In contrast, phages do not show any such pressure the remaining viruses, which mostly occur in plants and insects, do not show any significant pressure. All phages listed are ssRNA+.
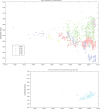
Figure 2. Comparison of CpG suppression in host and viral genomes.
Left: CpG odds ratio versus C+G content for human genes in blue. Superimposed on top of these genes are the human RNA viruses from Figure 1, in the same color scheme: Human ssRNA+ (black) and Human ssRNA−(red). Right: Natural logarithm of the CpG odds ratio (log(η)) versus C+G content for human (blue) and chicken (yellow) (Gallus gallus) coding genes of at least 500 bases. A value of log(η) close to 0 (marked in the figure with a dashed line), indicates that the observed number of CpGs is similar to the expected. CpGs preassures are stronger in human than in birds.
Figure 3. Evolution of the number of CpG motifs in influenza A and B.
Top: Evolution of the number of CpGs in influenza A virus in time. When an influenza virus crosses from birds to humans the virus mimics the host genes by reducing the number of CpGs. Bottom: Evolution of the number of CpGs in influenza B virus in time.
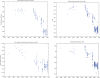
Figure 4. Different measures of dinucleotide suppression applied to the evolution of the human H1N1 virus.
Upper left figure: Evolution of the number of CpGs in human H1N1 influenza A virus from 1918–2007. Upper right figure: Evolution of the odds ratio η = CpG/C.G in human H1N1 influenza A virus from 1918–2007. Lower left figure: Evolution of the ratio (r) of CG Arginine (CGN) codons over the total number of arginines (CGN, AGA, AGG) in human H1N1 influenza A virus from 1918–2007. Lower right figure: Evolution of the ρ index in human H1N1 influenza A virus from 1918–2007.
Similar articles
- The dinucleotide composition of the Zika virus genome is shaped by conflicting evolutionary pressures in mammalian hosts and mosquito vectors.
Fros JJ, Visser I, Tang B, Yan K, Nakayama E, Visser TM, Koenraadt CJM, van Oers MM, Pijlman GP, Suhrbier A, Simmonds P. Fros JJ, et al. PLoS Biol. 2021 Apr 19;19(4):e3001201. doi: 10.1371/journal.pbio.3001201. eCollection 2021 Apr. PLoS Biol. 2021. PMID: 33872300 Free PMC article. - Does the Zinc Finger Antiviral Protein (ZAP) Shape the Evolution of Herpesvirus Genomes?
Lin YT, Chau LF, Coutts H, Mahmoudi M, Drampa V, Lee CH, Brown A, Hughes DJ, Grey F. Lin YT, et al. Viruses. 2021 Sep 17;13(9):1857. doi: 10.3390/v13091857. Viruses. 2021. PMID: 34578438 Free PMC article. Review. - Plant Virus Genome Is Shaped by Specific Dinucleotide Restrictions That Influence Viral Infection.
González de Prádena A, Sánchez Jimenez A, San León D, Simmonds P, García JA, Valli AA. González de Prádena A, et al. mBio. 2020 Feb 18;11(1):e02818-19. doi: 10.1128/mBio.02818-19. mBio. 2020. PMID: 32071264 Free PMC article. - Dinucleotide Composition in Animal RNA Viruses Is Shaped More by Virus Family than by Host Species.
Di Giallonardo F, Schlub TE, Shi M, Holmes EC. Di Giallonardo F, et al. J Virol. 2017 Mar 29;91(8):e02381-16. doi: 10.1128/JVI.02381-16. Print 2017 Apr 15. J Virol. 2017. PMID: 28148785 Free PMC article. - Evolution and ecology of influenza A viruses.
Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Webster RG, et al. Microbiol Rev. 1992 Mar;56(1):152-79. doi: 10.1128/mr.56.1.152-179.1992. Microbiol Rev. 1992. PMID: 1579108 Free PMC article. Review.
Cited by
- Selective Factors Associated with the Evolution of Codon Usage in Natural Populations of Arboviruses.
Velazquez-Salinas L, Zarate S, Eschbaumer M, Pereira Lobo F, Gladue DP, Arzt J, Novella IS, Rodriguez LL. Velazquez-Salinas L, et al. PLoS One. 2016 Jul 25;11(7):e0159943. doi: 10.1371/journal.pone.0159943. eCollection 2016. PLoS One. 2016. PMID: 27455096 Free PMC article. - The ecology and adaptive evolution of influenza A interspecies transmission.
Joseph U, Su YC, Vijaykrishna D, Smith GJ. Joseph U, et al. Influenza Other Respir Viruses. 2017 Jan;11(1):74-84. doi: 10.1111/irv.12412. Epub 2016 Aug 8. Influenza Other Respir Viruses. 2017. PMID: 27426214 Free PMC article. Review. - Origin of the 1918 pandemic H1N1 influenza A virus as studied by codon usage patterns and phylogenetic analysis.
Anhlan D, Grundmann N, Makalowski W, Ludwig S, Scholtissek C. Anhlan D, et al. RNA. 2011 Jan;17(1):64-73. doi: 10.1261/rna.2395211. Epub 2010 Nov 10. RNA. 2011. PMID: 21068184 Free PMC article. - Codon Usage Bias Analysis of Bluetongue Virus Causing Livestock Infection.
Yao X, Fan Q, Yao B, Lu P, Rahman SU, Chen D, Tao S. Yao X, et al. Front Microbiol. 2020 May 19;11:655. doi: 10.3389/fmicb.2020.00655. eCollection 2020. Front Microbiol. 2020. PMID: 32508755 Free PMC article. - Inferring the hosts of coronavirus using dual statistical models based on nucleotide composition.
Tang Q, Song Y, Shi M, Cheng Y, Zhang W, Xia XQ. Tang Q, et al. Sci Rep. 2015 Nov 26;5:17155. doi: 10.1038/srep17155. Sci Rep. 2015. PMID: 26607834 Free PMC article.
References
- Josse J, Kaiser AD, Kornberg A. Enzymatic Synthesis of Deoxyribonucleic Acid VIII. J Biol Chem. 1961;236:864–867. - PubMed
- Swartz MN, Trautner Tam Kornberg A. Enzymatic Synthesis of Deoxyribonucleic Acid XI. J Biol Chem. 1962;237:1961–1967. - PubMed
- Salser W. Globin mRNA sequences: analysis of base-pairing and evolutionary implications. Cold Spring Harbour Symp Quant Biol. 1977;42:985–1002. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials