B-cell lymphoma 6 and the molecular pathogenesis of diffuse large B-cell lymphoma - PubMed (original) (raw)
Review
B-cell lymphoma 6 and the molecular pathogenesis of diffuse large B-cell lymphoma
Weimin Ci et al. Curr Opin Hematol. 2008 Jul.
Abstract
Purpose of review: The B-cell lymphoma 6 transcriptional repressor is the most commonly involved oncogene in B-cell lymphomas. Sustained expression of B-cell lymphoma 6 causes malignant transformation of germinal center B cells. Understanding the mechanism of action of B-cell lymphoma 6 is crucial for the study of how aberrant transcriptional programming leads to lymphomagenesis and development of targeted antilymphoma therapy.
Recent findings: Identification of B-cell lymphoma 6 target genes indicates a critical role for B-cell lymphoma 6 in facilitating a state of physiological genomic instability required for germinal center B cells to undergo affinity maturation, and suggests its contribution to several additional cellular functions. The discovery of several layers of counterregulatory mechanisms reveals how B cells can control and fine-tune the potentially lymphomagenic actions of B-cell lymphoma 6. From the biochemical standpoint, B-cell lymphoma 6 can regulate distinct biological pathways through different cofactors. This observation explains how the biological actions of B-cell lymphoma 6 can be physiologically controlled through separate mechanisms and affords the means for improved therapeutic targeting. The fact that patients with B-cell lymphoma 6-dependent lymphoma can be identified on the basis of gene signatures suggests that therapeutic trials of B-cell lymphoma 6 inhibitors could be personalized to these individuals.
Summary: B-cell lymphoma 6 plays a fundamental role in lymphomagenesis and is an excellent therapeutic target for development of improved antilymphoma therapeutic regimens.
Figures
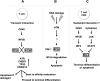
Figure 1. BCL6 and AID contribute to lymphomagenesis by facilitating genomic instability in GC B-cells
A: Naïve B-cells are triggered to form germinal centers by T-cell dependent immune responses. B: AID and BCL6 are upregulated as B-cells differentiate into centroblasts. AID mediates somatic hypermutation while BCL6 represses ATR and TP53 to attenuate DNA damage responses, and represses PRDM1 to block further differentiation. C: AID induces genetic lesions that lead to constitutive expression of BCL6, which in turn leads to sustained repression of ATR, TP53 and PRDM1. This forces B-cells to lock in to the GC B-cell phenotype and prevents them from undergoing terminal differentiation (D). E: The combined action of AID and BCL6 result in genomic instability and lymphomagenesis. F: Constitutive expression of BCL6 maintains DLBCL cell survival and may lead to further mutagenesis.
Figure 2. Inhibitory mechanisms for BCL6 function or expression
A: Reversible inhibition of BCL6 through transient CD40 signaling: CD40 signaling triggers NFkB activity, which results in dissociation of the BCL6-N-CoR complex and release of BCL6 repression of checkpoint genes. This allows cells to undergo a DNA damage sensing quality control step leading to apoptosis if damage is severe; or once CD40 signaling ceases, to undergo additional rounds of affinity maturation, or to proceed to terminal differentiation. B: ATM induced BCL6 protein degradation. High levels of DNA damage trigger an ATM dependent cascade that results in BCL6 phosphorylation and association with PIN1, followed by proteolytic degradation of BCL6 and apoptosis. C: CD40 mediated transcriptional downregulation of BCL6. Sustained contact between FDCs and B-cells leads to NFkB induction of IRF4, which can repress BCL6, while at the same time cytokines can trigger JAK-STAT pathways whereby STAT5 can also repress BCL6. This leads to terminal differentiation of selected high affinity immunoglobulin producing cells.
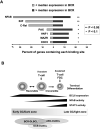
Figure 3. The balance of power between BCL6, NFkB and STAT3
A: A transcription factor binding site analysis (TRANSFAC) was performed within DNA sequences corresponding to the binding peak of BCL6 within target genes identified by ChIP-on-chip[27]. Light grey bars represent the percent of genes that are differentially expressed below the median in BCL6-dependent BCR-type (B-cell receptor signature) DLBCLs while the percent of genes above the median are represented in dark grey. The P value represents the significance of differential representation of genes above or below the median for each of the indicated transcription factor binding sites. B: BCL6 expression is upregulated when B-cells enter the germinal center reaction. In dark zone centroblasts BCL6 can block NFkB signaling through several mechanisms and directly repress STAT3 as described in the text. Transient interactions with T-cells can induce B-cell NFkB activity in a reversible fashion, which may allow B-cells to recycle back to the dark zone. Conversely, the actions of cytokines and/or more prolonged contact with FDCs or T-cells can tip the balance towards NFkB and STAT3 and downregulated BCL6 expression. GCB DLBCLs are generally proposed to reflect the biology of cells earlier in the germinal center reaction, while ABC DLBCLs are generally believed to represent later and/or exiting cells. The BCR/BCL6 dependent type of DLBCLs may partially overlap with both of these categories, but their BCL6 dependent biology suggests that they have features that overlap with GC B-cells in which BCL6 is functionally active.
Figure 4. BCL6 regulates different biological functions through distinct corepressors
A: The graphical representation depicts two molecules of BCL6 forming a homodimer through the BTB domain. The BCL6 BTB domain can recruit SMRT, N-CoR and BCoR through the lateral groove. Repression of ATR, CHEK1, TP53 and CDKN1A is dependent on these corepressors and mediates survival and proliferation during AID induced somatic hypermutation. A region of BCL6 proximal to the BTB domain recruits CtBP, which can mediate negative autoregulation of BCL6. The central second repression domain (RD2) of BCL6 recruits an MTA3/NuRD complex, which is required for repression of PRDM1 and thus mediates differentiation blockade. The zinc finger (ZF) domain can bind directly to ETO [52], although a specific biological function for this corepressor is not yet known. B: A gene ontology analysis of BCL6 target genes identified by ChIP-on-chip shows them to be associated with specific biological functions as indicated in the pie chart.
Similar articles
- BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms.
Parekh S, Polo JM, Shaknovich R, Juszczynski P, Lev P, Ranuncolo SM, Yin Y, Klein U, Cattoretti G, Dalla Favera R, Shipp MA, Melnick A. Parekh S, et al. Blood. 2007 Sep 15;110(6):2067-74. doi: 10.1182/blood-2007-01-069575. Epub 2007 Jun 1. Blood. 2007. PMID: 17545502 Free PMC article. - BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis.
Basso K, Dalla-Favera R. Basso K, et al. Adv Immunol. 2010;105:193-210. doi: 10.1016/S0065-2776(10)05007-8. Adv Immunol. 2010. PMID: 20510734 Review. - The CREBBP Acetyltransferase Is a Haploinsufficient Tumor Suppressor in B-cell Lymphoma.
Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, Meyer SN, Mo T, Basso K, Brindle PK, Hussein S, Dalla-Favera R, Pasqualucci L. Zhang J, et al. Cancer Discov. 2017 Mar;7(3):322-337. doi: 10.1158/2159-8290.CD-16-1417. Epub 2017 Jan 9. Cancer Discov. 2017. PMID: 28069569 Free PMC article. - BCL6 as a therapeutic target for lymphoma.
Leeman-Neill RJ, Bhagat G. Leeman-Neill RJ, et al. Expert Opin Ther Targets. 2018 Feb;22(2):143-152. doi: 10.1080/14728222.2018.1420782. Epub 2018 Jan 4. Expert Opin Ther Targets. 2018. PMID: 29262721 Review. - [Signaling pathways in pathogenesis of diffuse large B-cell lymphoma].
Zhang F, Xu FP, Liu YH, Zhuang HG. Zhang F, et al. Zhonghua Bing Li Xue Za Zhi. 2011 Apr;40(4):282-5. Zhonghua Bing Li Xue Za Zhi. 2011. PMID: 21616012 Review. Chinese. No abstract available.
Cited by
- Imatinib disrupts lymphoma angiogenesis by targeting vascular pericytes.
Ruan J, Luo M, Wang C, Fan L, Yang SN, Cardenas M, Geng H, Leonard JP, Melnick A, Cerchietti L, Hajjar KA. Ruan J, et al. Blood. 2013 Jun 27;121(26):5192-202. doi: 10.1182/blood-2013-03-490763. Epub 2013 Apr 30. Blood. 2013. PMID: 23632889 Free PMC article. - B-cell Lymphoma 6 (BCL6): From Master Regulator of Humoral Immunity to Oncogenic Driver in Pediatric Cancers.
McLachlan T, Matthews WC, Jackson ER, Staudt DE, Douglas AM, Findlay IJ, Persson ML, Duchatel RJ, Mannan A, Germon ZP, Dun MD. McLachlan T, et al. Mol Cancer Res. 2022 Dec 2;20(12):1711-1723. doi: 10.1158/1541-7786.MCR-22-0567. Mol Cancer Res. 2022. PMID: 36166198 Free PMC article. Review. - DNA methylation prevents CTCF-mediated silencing of the oncogene BCL6 in B cell lymphomas.
Lai AY, Fatemi M, Dhasarathy A, Malone C, Sobol SE, Geigerman C, Jaye DL, Mav D, Shah R, Li L, Wade PA. Lai AY, et al. J Exp Med. 2010 Aug 30;207(9):1939-50. doi: 10.1084/jem.20100204. Epub 2010 Aug 23. J Exp Med. 2010. PMID: 20733034 Free PMC article. - Wilms' tumor 1-associating protein plays an aggressive role in diffuse large B-cell lymphoma and forms a complex with BCL6 via Hsp90.
Kuai Y, Gong X, Ding L, Li F, Lei L, Gong Y, Liu Q, Tan H, Zhang X, Liu D, Ren G, Pan H, Shi Y, Berberich-Siebelt F, Mao Z, Zhou R. Kuai Y, et al. Cell Commun Signal. 2018 Aug 24;16(1):50. doi: 10.1186/s12964-018-0258-6. Cell Commun Signal. 2018. PMID: 30143009 Free PMC article. - The feedback loop of LITAF and BCL6 is involved in regulating apoptosis in B cell non-Hodgkin's-lymphoma.
Shi Y, Kuai Y, Lei L, Weng Y, Berberich-Siebelt F, Zhang X, Wang J, Zhou Y, Jiang X, Ren G, Pan H, Mao Z, Zhou R. Shi Y, et al. Oncotarget. 2016 Nov 22;7(47):77444-77456. doi: 10.18632/oncotarget.12680. Oncotarget. 2016. PMID: 27764808 Free PMC article.
References
- Allman D, Jain A, Dent A, et al. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. - PubMed
- Dent AL, Shaffer AL, Yu X, et al. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. - PubMed
- Ye BH, Cattoretti G, Shen Q, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. - PubMed
- Ye BH. BCL-6 in the pathogenesis of non-Hodgkin's lymphoma. Cancer Invest. 2000;18:356–365. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials