Image processing for electron microscopy single-particle analysis using XMIPP - PubMed (original) (raw)
Image processing for electron microscopy single-particle analysis using XMIPP
Sjors H W Scheres et al. Nat Protoc. 2008.
Abstract
We describe a collection of standardized image processing protocols for electron microscopy single-particle analysis using the XMIPP software package. These protocols allow performing the entire processing workflow starting from digitized micrographs up to the final refinement and evaluation of 3D models. A particular emphasis has been placed on the treatment of structurally heterogeneous data through maximum-likelihood refinements and self-organizing maps as well as the generation of initial 3D models for such data sets through random conical tilt reconstruction methods. All protocols presented have been implemented as stand-alone, executable python scripts, for which a dedicated graphical user interface has been developed. Thereby, they may provide novice users with a convenient tool to quickly obtain useful results with minimum efforts in learning about the details of this comprehensive package. Examples of applications are presented for a negative stain random conical tilt data set on the hexameric helicase G40P and for a structurally heterogeneous data set on 70S Escherichia coli ribosomes embedded in vitrified ice.
Figures
Figure 1
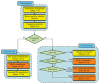
A generalized XMIPP processing workflow. The protocols developed may be divided in data preprocessing, 2D processing and 3D processing (light-blue boxes). Computationally demanding protocols that allow multiprocessor computing (via message-passing interface, MPI) are shown in orange; all other protocols are shown in yellow.
Figure 2
Micrograph selection. This is based on (a–c) CTFs and on (d–f) particle appearance. (a) A suitable CTF has several rotationally symmetric rings. CTFs should be discarded if they present drift, that is, (b) fading in a particular direction, or astigmatism, that is, (c) rotationally asymmetric. Furthermore, suitable micrographs should present a (d) homogenous population of well-separated particles. (e) Micrographs should be discarded if the particle density is too high, that is, the particles are so close to each other that they almost overlap, or (f) if the particles are very heterogeneous in size or appearance, indicating aggregation or other forms of particle instability. Panels d–f are on the same scale, and the scale bar in panel d represents 50 nm.
Figure 3
Example of class selection from a self-organizing map of rotational spectra. The user interactively identifies distinct classes, each of which may comprehend several (neighboring) code vectors. In this example, two classes were identified, one with sixfold symmetry and one with threefold symmetry. Code vectors were identified as belonging to the sixfold symmetric class when they contained a single peak at the sixfold harmonic. Code vectors were identified to belong to the threefold symmetric class when they contained the largest peak at the threefold harmonic and a secondary peak at the sixfold harmonic. The graphical interface of the
xmipp_show
program (Steps 14 and 20) allows calculating averages for all experimental measurements corresponding to the selected classes (insets).
Figure 4
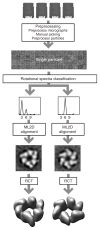
Anticipated results for the G40P case. Tilted pairs of digitized micrographs were processed to yield a data set of 14,000 particle pairs. The untilted particles were classified in threefold and sixfold symmetric particles based on their rotational spectra. For both classes, the untilted particles were aligned using maximum-likelihood refinement, and the tilted particles were used to calculate the corresponding random conical tilt reconstructions.
Figure 5
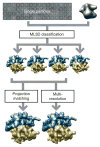
Anticipated results for the ribosome case. A structurally heterogeneous data set of 70S E. coli ribosome particles in complex with or without EFG were classified using ML3D classification, and the corresponding class with particles lacking EFG was further refined using either projection matching or multiresolution refinement.
Similar articles
- Xmipp 3.0: an improved software suite for image processing in electron microscopy.
de la Rosa-Trevín JM, Otón J, Marabini R, Zaldívar A, Vargas J, Carazo JM, Sorzano CO. de la Rosa-Trevín JM, et al. J Struct Biol. 2013 Nov;184(2):321-8. doi: 10.1016/j.jsb.2013.09.015. Epub 2013 Sep 26. J Struct Biol. 2013. PMID: 24075951 - Hybrid Electron Microscopy Normal Mode Analysis graphical interface and protocol.
Sorzano CO, de la Rosa-Trevín JM, Tama F, Jonić S. Sorzano CO, et al. J Struct Biol. 2014 Nov;188(2):134-41. doi: 10.1016/j.jsb.2014.09.005. Epub 2014 Sep 27. J Struct Biol. 2014. PMID: 25268657 - XMIPP: a new generation of an open-source image processing package for electron microscopy.
Sorzano CO, Marabini R, Velázquez-Muriel J, Bilbao-Castro JR, Scheres SH, Carazo JM, Pascual-Montano A. Sorzano CO, et al. J Struct Biol. 2004 Nov;148(2):194-204. doi: 10.1016/j.jsb.2004.06.006. J Struct Biol. 2004. PMID: 15477099 - Three-dimensional reconstruction of single particles from random and nonrandom tilt series.
Radermacher M. Radermacher M. J Electron Microsc Tech. 1988 Aug;9(4):359-94. doi: 10.1002/jemt.1060090405. J Electron Microsc Tech. 1988. PMID: 3058896 Review. - Single-Particle Refinement and Variability Analysis in EMAN2.1.
Ludtke SJ. Ludtke SJ. Methods Enzymol. 2016;579:159-89. doi: 10.1016/bs.mie.2016.05.001. Epub 2016 Jul 1. Methods Enzymol. 2016. PMID: 27572727 Free PMC article. Review.
Cited by
- Prevention of overfitting in cryo-EM structure determination.
Scheres SH, Chen S. Scheres SH, et al. Nat Methods. 2012 Sep;9(9):853-4. doi: 10.1038/nmeth.2115. Nat Methods. 2012. PMID: 22842542 Free PMC article. No abstract available. - Identification of a Membrane-bound Prepore Species Clarifies the Lytic Mechanism of Actinoporins.
Morante K, Bellomio A, Gil-Cartón D, Redondo-Morata L, Sot J, Scheuring S, Valle M, González-Mañas JM, Tsumoto K, Caaveiro JMM. Morante K, et al. J Biol Chem. 2016 Sep 9;291(37):19210-19219. doi: 10.1074/jbc.M116.734053. Epub 2016 Jul 21. J Biol Chem. 2016. PMID: 27445331 Free PMC article. - Data management challenges in three-dimensional EM.
Patwardhan A, Carazo JM, Carragher B, Henderson R, Heymann JB, Hill E, Jensen GJ, Lagerstedt I, Lawson CL, Ludtke SJ, Mastronarde D, Moore WJ, Roseman A, Rosenthal P, Sorzano CO, Sanz-García E, Scheres SH, Subramaniam S, Westbrook J, Winn M, Swedlow JR, Kleywegt GJ. Patwardhan A, et al. Nat Struct Mol Biol. 2012 Dec;19(12):1203-7. doi: 10.1038/nsmb.2426. Nat Struct Mol Biol. 2012. PMID: 23211764 Free PMC article. - Conformational transitions regulate the exposure of a DNA-binding domain in the RuvBL1-RuvBL2 complex.
López-Perrote A, Muñoz-Hernández H, Gil D, Llorca O. López-Perrote A, et al. Nucleic Acids Res. 2012 Nov;40(21):11086-99. doi: 10.1093/nar/gks871. Epub 2012 Sep 21. Nucleic Acids Res. 2012. PMID: 23002137 Free PMC article. - The atypical subunit composition of respiratory complexes I and IV is associated with original extra structural domains in Euglena gracilis.
Miranda-Astudillo HV, Yadav KNS, Colina-Tenorio L, Bouillenne F, Degand H, Morsomme P, Boekema EJ, Cardol P. Miranda-Astudillo HV, et al. Sci Rep. 2018 Jun 26;8(1):9698. doi: 10.1038/s41598-018-28039-z. Sci Rep. 2018. PMID: 29946152 Free PMC article.
References
- Henderson R. Realizing the potential of electron cryo-microscopy. Q Rev Biophys. 2004;37:3–13. - PubMed
- Subramaniam S, Milne JLS. Three-dimensional electron microscopy at molecular resolution. Annu Rev Biophys Biomol Struct. 2004;33:141–155. - PubMed
- Carragher B, Smith PR. Special issue on Advances in Computational Image Processing for Microscopy. J Struct Biol. 1996;116:2–8. - PubMed
- Chiu SLAW. Special Issue on Single Particle Processing. In: Ludtke SJ, Chiu W, editors. J Struct Biol. Vol. 173. 2001.
- Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources