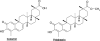Natural compounds with proteasome inhibitory activity for cancer prevention and treatment - PubMed (original) (raw)
Review
Natural compounds with proteasome inhibitory activity for cancer prevention and treatment
H Yang et al. Curr Protein Pept Sci. 2008 Jun.
Abstract
The proteasome is a multicatalytic protease complex that degrades most endogenous proteins including misfolded or damaged proteins to ensure normal cellular function. The ubiquitin-proteasome degradation pathway plays an essential role in multiple cellular processes, including cell cycle progression, proliferation, apoptosis and angiogenesis. It has been shown that human cancer cells are more sensitive to proteasome inhibition than normal cells, indicating that a proteasome inhibitor could be used as a novel anticancer drug. Indeed, this idea has been supported by the encouraging results of the clinical trials using the proteasome inhibitor Bortezomib (Velcade, PS-341), a drug approved by the US Food and Drug Administration (FDA). Several natural compounds, including the microbial metabolite lactacystin, green tea polyphenols, and traditional medicinal triterpenes, have been shown to be potent proteasome inhibitors. These findings suggest the potential use of natural proteasome inhibitors as not only chemopreventive and chemotherapeutic agents, but also tumor sensitizers to conventional radiotherapy and chemotherapy. In this review, we will summarize the structures and biological activities of the proteasome and several natural compounds with proteasome inhibitory activity, and will discuss the potential use of these compounds for the prevention and treatment of human cancers.
Figures
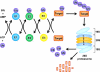
Fig. (1). The Ubiquitin-Proteasome Pathway
Ubiquitin (Ub) is activated by Ub-activating (E1) enzymes through adenylation and formation of high-energy thiol ester bond and then transferred to Ub-conjugating (E2) enzymes. With the help of Ub-ligating (E3) enzymes, Ub is finally tranferred to a reactive lysine residue of a target protein. Ubiquitinated proteins are recognized by the 19S cap of the 26S proteasome and fed into its 20S catalytic core for degradation into oligopeptides. The Ub is then released and recycled.
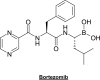
Fig. (2)
Chemical structure of Bortezomib.
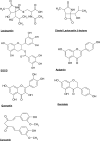
Fig. (3)
Chemical structures of lactacystin, clasto-lactacystin β-lactone and several natural polyphenols.
Fig. (4)
Chemical structures of natural triterpenes.
Similar articles
- Targeting the proteasome pathway.
Tsukamoto S, Yokosawa H. Tsukamoto S, et al. Expert Opin Ther Targets. 2009 May;13(5):605-21. doi: 10.1517/14728220902866851. Expert Opin Ther Targets. 2009. PMID: 19397479 Review. - [Proteasome inhibitor].
Yamamura M, Hirai T, Yamaguchi Y. Yamamura M, et al. Nihon Rinsho. 2010 Jun;68(6):1079-84. Nihon Rinsho. 2010. PMID: 20535959 Review. Japanese. - The proteasome as a potential target for novel anticancer drugs and chemosensitizers.
Landis-Piwowar KR, Milacic V, Chen D, Yang H, Zhao Y, Chan TH, Yan B, Dou QP. Landis-Piwowar KR, et al. Drug Resist Updat. 2006 Dec;9(6):263-73. doi: 10.1016/j.drup.2006.11.001. Epub 2007 Jan 2. Drug Resist Updat. 2006. PMID: 17197231 Review. - The proteasome: a novel target for anticancer therapy.
Montagut C, Rovira A, Albanell J. Montagut C, et al. Clin Transl Oncol. 2006 May;8(5):313-7. doi: 10.1007/s12094-006-0176-8. Clin Transl Oncol. 2006. PMID: 16760005 Review. - Proteasome: an emerging target for cancer therapy.
Zavrski I, Jakob C, Schmid P, Krebbel H, Kaiser M, Fleissner C, Rosche M, Possinger K, Sezer O. Zavrski I, et al. Anticancer Drugs. 2005 Jun;16(5):475-81. doi: 10.1097/00001813-200506000-00002. Anticancer Drugs. 2005. PMID: 15846112 Review.
Cited by
- Chalcone-based small-molecule inhibitors attenuate malignant phenotype via targeting deubiquitinating enzymes.
Issaenko OA, Amerik AY. Issaenko OA, et al. Cell Cycle. 2012 May 1;11(9):1804-17. doi: 10.4161/cc.20174. Epub 2012 May 1. Cell Cycle. 2012. PMID: 22510564 Free PMC article. - Proteasome inhibition mediates p53 reactivation and anti-cancer activity of 6-gingerol in cervical cancer cells.
Rastogi N, Duggal S, Singh SK, Porwal K, Srivastava VK, Maurya R, Bhatt ML, Mishra DP. Rastogi N, et al. Oncotarget. 2015 Dec 22;6(41):43310-25. doi: 10.18632/oncotarget.6383. Oncotarget. 2015. PMID: 26621832 Free PMC article. - Celastrol and Its Role in Controlling Chronic Diseases.
Venkatesha SH, Moudgil KD. Venkatesha SH, et al. Adv Exp Med Biol. 2016;928:267-289. doi: 10.1007/978-3-319-41334-1_12. Adv Exp Med Biol. 2016. PMID: 27671821 Free PMC article. - Quercetin as a potential modulator of P-glycoprotein expression and function in cells of human pancreatic carcinoma line resistant to daunorubicin.
Borska S, Sopel M, Chmielewska M, Zabel M, Dziegiel P. Borska S, et al. Molecules. 2010 Feb 9;15(2):857-70. doi: 10.3390/molecules15020857. Molecules. 2010. PMID: 20335952 Free PMC article. - Thermally targeted p21 peptide enhances bortezomib cytotoxicity in androgen-independent prostate cancer cell lines.
Mikecin AM, Walker LR, Kuna M, Raucher D. Mikecin AM, et al. Anticancer Drugs. 2014 Feb;25(2):189-99. doi: 10.1097/CAD.0000000000000036. Anticancer Drugs. 2014. PMID: 24113592 Free PMC article.
References
- Lu M, Dou QP, Kitson RP, Smith DM, Goldfarb RH. J Cell Biochem. 2006;97:122–134. - PubMed
- Adams J, behnke Mark., Chen s., Cruickshank AA, Dick LR, Grenier L, Klunder JM, Ma YT, Plamondon L, Stein RL. Bioorg Med Chem Lett. 1998;8:333–338. - PubMed
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Nature. 1997;386:463–471. - PubMed
- Pickart CM, Cohen RE. Nat Rev Mol Cell Biol. 2004;5:177–187. - PubMed
- Nandi D, Tahiliani P, Kumar A, Chandu D. J Biosci. 2006;31:137–155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA120009-03/CA/NCI NIH HHS/United States
- R01 CA120009-04/CA/NCI NIH HHS/United States
- CA112625/CA/NCI NIH HHS/United States
- CA120009/CA/NCI NIH HHS/United States
- P30 CA022453/CA/NCI NIH HHS/United States
- R01 CA120009/CA/NCI NIH HHS/United States
- R03 CA112625/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources