GOLPH2 and MYO6: putative prostate cancer markers localized to the Golgi apparatus - PubMed (original) (raw)
GOLPH2 and MYO6: putative prostate cancer markers localized to the Golgi apparatus
Shuanzeng Wei et al. Prostate. 2008.
Abstract
Background: Malignant transformation is often accompanied by morphological and functional alterations in subcellular organelles. The Golgi apparatus is a subcellular structure primarily involved in modification and sorting of macromolecules for secretion and transport to other cellular destinations. Molecular alterations associated with the Golgi apparatus may take place during prostate carcinogenesis but such alterations have not been documented.
Methods: To demonstrate that the Golgi apparatus undergoes alterations during prostate carcinogenesis, we examined the expression and localization of two candidate molecules, Golgi phosphoprotein 2 (GOLPH2) and myosin VI (MYO6), both overexpressed in prostate cancer as initially identified by expression microarray analysis.
Results: Elevated GOLPH2 expression in prostate cancers was validated through real-time RT-PCR, Western blot, and tissue microarray analysis, and its Golgi localization in surgical prostate cancer tissues confirmed using two-color immunofluorescence. In addition, distinctive juxtanuclear MYO6 staining pattern consistent with Golgi localization was observed in surgical prostate cancer tissues. Two-color immunofluorescence revealed intensive Golgi-specific staining for both GOLPH2 and myosin VI in prostate cancer cells but not in the adjacent normal prostate epithelium.
Conclusions: We show that the Golgi apparatus in prostate cancer cells differs from the normal Golgi by elevated levels of two molecules, GOLPH2 and MYO6. These results for the first time demonstrated consistent cancer cell-specific alterations in the molecular composition of the Golgi apparatus. Such alterations can be explored for discovery of novel prostate cancer biomarkers through targeted organellar approaches.
(c) 2008 Wiley-Liss, Inc.
Figures
Fig. 1
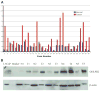
Validation of GOLPH2 mRNA and protein overexpression in human prostate cancer. A: Real-time RT-PCR analysis of GOLPH2 mRNA in prostate cancer tissues (n =30) and paired normal prostate tissues (n =30) from surgical prostate cancer specimens (_X_-axis). Normalized (against GAPDH) Ct values were converted to fold expression changes (_Y_-axis) relative to the median value of the normal samples. B: Western blot analysis of GOLPH2 protein in five paired normal/cancer prostate tissue samples collected fresh from five surgical cases. β-Actin is a loading control. LNCaP cell lysate was used as a positive control. N1–N5: normal prostate epithelial tissues, T1–T5: prostate cancer tissues.
Fig. 2
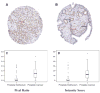
Tissue microarray (TMA) analysis of GOLPH2 expression in human prostate cancer. A: TMA spot containing predominantly Gleason 4 (red arrow) prostate cancer tissue. B: TMA spot containing mixed Gleason 3 (blue arrow) and Gleason 5 (red arrow) prostate cancer tissues and normal prostate epithelium (black arrow). Note that only cancer cells are positive for GOLPH2 and the staining pattern is consistent with Golgi localization. C,D: Box plots showing automated TMA image analysis results for GOLPH2 staining in normal and prostate cancer tissues. Altogether 132 normal and 93 cancer lesions were evaluated excluding array spots with poor quality and ambiguous diagnoses. Box plots showed differences in positively stained areas (by pixel ratios, C) and intensity scores (D) between the normal and cancerous prostate epithelium. Note that pixel ratios (see Materials and Methods Section) cannot reach high percentages due to Golgi-confined staining for GOLPH2. Each box is lined at lower quartile, median, and upper quartile values for each group. The whiskers show the 1.5× inter-quartile range of the data. The circles and asterisks indicate outliers and extreme outliers beyond the whiskers.
Fig. 3
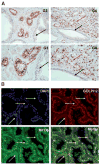
Immunohistochemical analysis of GOLPH2 and TGN46 in human prostate cancer tissues. A: Two adjacent serial sections of surgical prostate cancer tissues stained with GOLPH2 and TGN46, respectively, as indicated. Black arrows indicate cancer tissues, blue arrows indicate a normal prostate gland. Insets show high power view of the boxed areas. Note strong GOLPH2 staining in cancer cells and weak or negative staining in the normal prostate epithelium, and no difference for TGN46 staining in normal and cancer cells. B: Immunofluorescence double staining for TGN46 and GOLPH2 in the same formalin fixed paraffin embedded surgical prostate cancer section. Prostate cancer tissues were stained for nuclei (blue, by DAPI staining), TGN46 (green), GOLPH2 (red), and the images merged, as indicated. Note the nearly identical, relatively weak TGN46 staining in both normal and cancer cells, and cancer cell-specific overexpression of GOLPH2. White arrow indicates a normal duct and yellow arrows indicate cancerous acini.
Fig. 4
Myosin VI and GOLPH2 both localize to the Golgi apparatus. A: Prostate cancer tissues of varying histological grade (as indicated) stained with the myosin VI polyclonal antibody. Higher power views of a restricted area in top panels were presented immediately below. Note the distinctive perinuclear staining pattern present in the cancer epithelium (red arrows) regardless of the Gleason grades, and weak or absent staining in the normal prostate epithelium (black arrows). G3: Gleason 3; G4: predominantly Gleason 4. B: Myosin VI and GOLPH2 Immunofluorescence double labeling in formalin fixed paraffin embedded surgical prostate cancer tissues. Prostate cancer tissues were stained for nuclei (blue, by DAPI staining), MYO6 (green), GOLPH2 (red), and the images merged, as indicated. Note the distinctive Golgi staining pattern for GOLPH2 and MYO6 specifically in the cancer epithelium (yellow arrow), and the negative or weak signals for both proteins in the normal appearing prostate epithelium (white arrow).
Similar articles
- GOLPH2 protein expression as a novel tissue biomarker for prostate cancer: implications for tissue-based diagnostics.
Kristiansen G, Fritzsche FR, Wassermann K, Jäger C, Tölls A, Lein M, Stephan C, Jung K, Pilarsky C, Dietel M, Moch H. Kristiansen G, et al. Br J Cancer. 2008 Sep 16;99(6):939-48. doi: 10.1038/sj.bjc.6604614. Br J Cancer. 2008. PMID: 18781151 Free PMC article. - Golgi phosphoprotein 2 (GOLPH2) is a novel bile acid-responsive modulator of oesophageal cell migration and invasion.
Byrne AM, Bekiaris S, Duggan G, Prichard D, Kirca M, Finn S, Reynolds JV, Kelleher D, Long A. Byrne AM, et al. Br J Cancer. 2015 Nov 3;113(9):1332-42. doi: 10.1038/bjc.2015.350. Epub 2015 Oct 13. Br J Cancer. 2015. PMID: 26461057 Free PMC article. - Golgi phosphoprotein 2 (GOLPH2/GP73/GOLM1) interacts with secretory clusterin.
Zhou Y, Li L, Hu L, Peng T. Zhou Y, et al. Mol Biol Rep. 2011 Mar;38(3):1457-62. doi: 10.1007/s11033-010-0251-7. Epub 2010 Sep 15. Mol Biol Rep. 2011. PMID: 20842452 - Implications of the Golgi apparatus in prostate cancer.
Migita T, Inoue S. Migita T, et al. Int J Biochem Cell Biol. 2012 Nov;44(11):1872-6. doi: 10.1016/j.biocel.2012.06.004. Epub 2012 Jun 19. Int J Biochem Cell Biol. 2012. PMID: 22721754 Review. - Novel biomarkers and therapeutic targets for prostate cancer.
Williams RM, Naz RK. Williams RM, et al. Front Biosci (Schol Ed). 2010 Jan 1;2(2):677-84. doi: 10.2741/s93. Front Biosci (Schol Ed). 2010. PMID: 20036976 Review.
Cited by
- GOLPH2 expression may serve as diagnostic marker in seminomas.
Fritzsche FR, Kristiansen G, Riener MO, Dietel M, Oelrich B. Fritzsche FR, et al. BMC Urol. 2010 Feb 25;10:4. doi: 10.1186/1471-2490-10-4. BMC Urol. 2010. PMID: 20184749 Free PMC article. - Multiplexed Electrochemical Cancer Diagnostics With Automated Microfluidics.
Mercer C, Jones A, Rusling JF, Leech D. Mercer C, et al. Electroanalysis. 2019 Feb;31(2):208-211. doi: 10.1002/elan.201800632. Epub 2018 Nov 27. Electroanalysis. 2019. PMID: 32390709 Free PMC article. - A Systems Approach to Interrogate Gene Expression Patterns in African American Men Presenting with Clinically Localized Prostate Cancer.
Hardiman G, Savage SJ, Hazard ES, da Silveira WA, Morgan R, Harris A, Jefferson MS, Wilson RC, Caulder S, Ambrose L, Frey L, Wolf B, Gattoni-Celli S, Hughes Halbert C. Hardiman G, et al. Cancers (Basel). 2021 Oct 14;13(20):5143. doi: 10.3390/cancers13205143. Cancers (Basel). 2021. PMID: 34680291 Free PMC article. - Evaluation of ERG responsive proteome in prostate cancer.
Tan SH, Furusato B, Fang X, He F, Mohamed AA, Griner NB, Sood K, Saxena S, Katta S, Young D, Chen Y, Sreenath T, Petrovics G, Dobi A, McLeod DG, Sesterhenn IA, Saxena S, Srivastava S. Tan SH, et al. Prostate. 2014 Jan;74(1):70-89. doi: 10.1002/pros.22731. Epub 2013 Sep 21. Prostate. 2014. PMID: 24115221 Free PMC article. - Golgi phosphoprotein 2 in physiology and in diseases.
Kim HJ, Lv D, Zhang Y, Peng T, Ma X. Kim HJ, et al. Cell Biosci. 2012 Sep 10;2(1):31. doi: 10.1186/2045-3701-2-31. Cell Biosci. 2012. PMID: 22958594 Free PMC article.
References
- Marsh BJ, Howell KE. The mammalian Golgi complex debates. Nat Rev Mol Cell Biol. 2002;3:789–795. - PubMed
- Sütterlin C, Hsu P, Mallabiabarrena A, Malhotra V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109:359–369. - PubMed
- Colanzi A, Suetterlin C, Malhotra V. Cell-cycle-specific Golgi fragmentation: How and why? Curr Opin Cell Biol. 2003;15:462–467. - PubMed
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, Yates JR, III, Maiato H, Khodjakov A, Akhmanova A, Kaverina I. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. - PMC - PubMed
- Liu Z, Vong QP, Zheng Y. CLASPing microtubules at the trans-Golgi network. Dev Cell. 2007;12:839–840. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous