Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells - PubMed (original) (raw)
Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells
Jason Q Boone et al. Dev Neurobiol. 2008 Aug.
Abstract
Mammalian neural stem cells generate transit amplifying progenitors that expand the neuronal population, but these type of progenitors have not been studied in Drosophila. The Drosophila larval brain contains approximately 100 neural stem cells (neuroblasts) per brain lobe, which are thought to bud off smaller ganglion mother cells (GMCs) that each produce two post-mitotic neurons. Here, we use molecular markers and clonal analysis to identify a novel neuroblast cell lineage containing "transit amplifying GMCs" (TA-GMCs). TA-GMCs differ from canonical GMCs in several ways: each TA-GMC has nuclear Deadpan, cytoplasmic Prospero, forms Prospero crescents at mitosis, and generates up to 10 neurons; canonical GMCs lack Deadpan, have nuclear Prospero, lack Prospero crescents at mitosis, and generate two neurons. We conclude that there are at least two types of neuroblast lineages: a Type I lineage where GMCs generate two neurons, and a type II lineage where TA-GMCs have longer lineages. Type II lineages allow more neurons to be produced faster than Type I lineages, which may be advantageous in a rapidly developing organism like Drosophila.
(c) 2008 Wiley Periodicals, Inc. Develop Neurobiol, 2008.
Figures
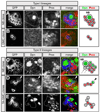
Figure 1. Clonal analysis identifies two types of larval neuroblast lineages
(A–E) Neuroblast (NB) and GMC clones stained for Deadpan (Dpn, green), Prospero (Pros, red), and the clone marker GFP (green, outlined). Right panel shows summary of markers: green, nuclear Dpn cytoplasmic Pros (Dpn+ Proscyto); red, Dpn-negative nuclear Pros (Dpn− Prosnucl). Type I clones were assayed in the DAL brain region; type II clones were assayed in the DPM brain region. (A) Type I neuroblast clone containing one large Dpn+ Proscyto NB and many Dpn− Prosnucl GMCs. (B) Type I GMC clone containing two Prosnucl immature neurons that lack GFP+ axons (data not shown). (C) Type II neuroblast clone containing one large Dpn+ Proscyto NB and smaller progeny including two Dpn− Proscyto cells closely-associated with the neuroblast (arrows), several Dpn+ Proscyto cells, and several Dpn− Prosnucl cells. (D) Type II TA-GMC small clone containing one Dpn+ Proscyto cell (arrowhead) and three Dpn− Prosnucl cells. (E) Type II TA-GMC large clone containing several Dpn− Prosnucl cells, and a pool of Dpn− Pros− mature neurons (based on their GFP+ axon projections). Scale bar = 6.24 µm.
Figure 2. Type I and type II neuroblast lineage locations within the brain
(A–C) Schematics of the third instar larval brain showing brain regions according to Pereanu and Hartenstein (2006). Type I neuroblast are found in all brain regions, but only type I neuroblasts are located in the dorsoanterior lateral (DAL) region, which is where we performed all type I lineage assays. Type II neuroblast are found in several brain regions (yellow shading); the largest number are in the dorsoposterior medial (DPM) region. The orientation of each brain is indicated by the axial arrows. OL, optic lobe. (A'–B') Three-dimensional reconstruction of a confocal image stack of a brain lobe double-label for Dpn (green spheres, neuroblasts; silver spheres, TA-GMCs) and Fasciculin II (mushroom body, red). Orientation is the same as the panel above. This brain lobe is shown in Movie 2. (C') Three-dimensional reconstruction of a confocal image stack of a brain lobe containing a type II neuroblast clone (white) stained for Fasciculin II (mushroom body lobes, red). The clone extends medially across the midline but the fine axon processes do not show up in this image; they can be seen in Movie 1. (C' inset) Optical section from brain used to generate the image shown panel C'. Clone marker (GFP, white); Deadpan (red); neuroblast in clone, arrow; TA-GMCs in clone, bracket. Abbreviations: OL, optic lobe; DPM, dorsoposterior medial (yellow outline); DAL, dorsoanterior lateral; DAM, dorsoanterior medial; DPL, dorsoposterior lateral; BLP, basolateral posterior; BLA, basolateral anterior; BLD, basolateral dorsal; BA, basoanterior; CP, centroposterior; CM, centromedial. Regions in smaller fonts are towards the back of the lobe. Scale bar = 20 µm.
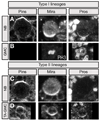
Figure 3. Asymmetric cell division within type I and type II neuroblast lineages
Mitotic larval neuroblasts and GMCs stained for the apical protein Partner of Inscuteable (Pins) and the basal proteins Miranda (Mira) and Prospero (Pros), in some cases the mitotic marker phosphohistone H3 (PH3) is shown. Pros/Mira or Pros/PH3 panels always show the same neuroblast. (A) Type I mitotic neuroblast in the DAL region shows apical Pins and basal Mira/Pros. Pros/Mira panels show the same NB. (B) Type I mitotic GMC in the DAL region shows diffuse cytoplasmic Pros (bright punctate staining in the Pros panel is DNA-associated Pros protein). (C) Type II mitotic neuroblast in the DPM region identified by lack of asymmetric Pros localization, despite completely normal localization of Pins and Mira. Pros/Mira panels show the same NB. (D) Type II mitotic TA-GMCs in the DPM identified by their small size and asymmetric localization of Pros and Mira to the cortex opposite the Pins. Scale bar = 6.24 µm.
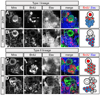
Figure 4. BrdU pulse/chase analysis of type I and type II neuroblast lineages
Larvae were pulsed with BrdU for 4.5h and then either fixed immediately ("pulse"; A, C) or grown without BrdU for 18h before fixing ("chase"; B, D). Larvae were stained for Miranda (Mira, green), BrdU (red), and the neuronal marker Elav (blue); neuroblasts (NBs), arrows; neuroblast progeny, brackets; schematics are shown to the right. Type I lineage data were collected in the DAL brain region; type II lineage data were collected in the DPM brain region. (A–B) Type I neuroblasts always incorporate BrdU during the pulse and dilute it out during the chase, whereas only a few type I GMCs contacting the neuroblast incorporate BrdU during the pulse (A, brackets); following the chase, BrdU is maintained in Elav+ post-mitotic neurons (B, brackets). (C–D) Type II neuroblasts always incorporate BrdU during the pulse and dilute it out during the chase; many type II progeny incorporate BrdU during the pulse (C, brackets); following the chase, BrdU is maintained in Elav+ post-mitotic neurons (D, brackets; shown in an inset, because the neurons are at the bottom of this confocal image stack). Scale bar = 6.24 µm.
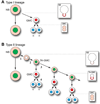
Figure 5. Summary of type I and type II larval neuroblast lineages
(A) Proposed type I neuroblast lineage. Nuclear Dpn (green), nuclear Pros (red), cytoplasmic or undetectable Pros (light red), cortical Prospero (red crescent), neuronal marker Elav (blue). Mitotic profiles are shown in boxes at right. See text for details. (B) Proposed type II neuroblast lineage. Nuclear Dpn (green), nuclear Pros (red), cytoplasmic or undetectable Pros (light red), cortical Prospero (red crescent), weak or undetectable cortical Pros (dashed red crescent), neuronal marker Elav (blue). Mitotic profiles are shown in boxes at right. See Discussion for details of each numbered step in the lineage.
Similar articles
- Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development.
Bello BC, Izergina N, Caussinus E, Reichert H. Bello BC, et al. Neural Dev. 2008 Feb 19;3:5. doi: 10.1186/1749-8104-3-5. Neural Dev. 2008. PMID: 18284664 Free PMC article. - Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex.
Bayraktar OA, Boone JQ, Drummond ML, Doe CQ. Bayraktar OA, et al. Neural Dev. 2010 Oct 1;5:26. doi: 10.1186/1749-8104-5-26. Neural Dev. 2010. PMID: 20920301 Free PMC article. - Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions.
Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Ikeshima-Kataoka H, et al. Nature. 1997 Dec 11;390(6660):625-9. doi: 10.1038/37641. Nature. 1997. PMID: 9403694 - Generating neuronal diversity in the Drosophila central nervous system: a view from the ganglion mother cells.
Karcavich RE. Karcavich RE. Dev Dyn. 2005 Mar;232(3):609-16. doi: 10.1002/dvdy.20273. Dev Dyn. 2005. PMID: 15704126 Review. - The development of the Drosophila larval brain.
Hartenstein V, Spindler S, Pereanu W, Fung S. Hartenstein V, et al. Adv Exp Med Biol. 2008;628:1-31. doi: 10.1007/978-0-387-78261-4_1. Adv Exp Med Biol. 2008. PMID: 18683635 Review.
Cited by
- Asymmetric cell division of stem and progenitor cells during homeostasis and cancer.
Gómez-López S, Lerner RG, Petritsch C. Gómez-López S, et al. Cell Mol Life Sci. 2014 Feb;71(4):575-97. doi: 10.1007/s00018-013-1386-1. Epub 2013 Jun 15. Cell Mol Life Sci. 2014. PMID: 23771628 Free PMC article. Review. - Seven-up acts in neuroblasts to specify adult central complex neuron identity and initiate neuroblast decommissioning.
Dillon NR, Manning L, Hirono K, Doe CQ. Dillon NR, et al. Development. 2024 Feb 1;151(3):dev202504. doi: 10.1242/dev.202504. Epub 2024 Feb 1. Development. 2024. PMID: 38230563 Free PMC article. - Analysis of cell identity, morphology, apoptosis and mitotic activity in a primary neural cell culture system in Drosophila.
Moraru MM, Egger B, Bao DB, Sprecher SG. Moraru MM, et al. Neural Dev. 2012 Jun 20;7:14. doi: 10.1186/1749-8104-7-14. Neural Dev. 2012. PMID: 22554060 Free PMC article. - Drosophila ClC-a is required in glia of the stem cell niche for proper neurogenesis and wiring of neural circuits.
Plazaola-Sasieta H, Zhu Q, Gaitán-Peñas H, Rios M, Estévez R, Morey M. Plazaola-Sasieta H, et al. Glia. 2019 Dec;67(12):2374-2398. doi: 10.1002/glia.23691. Epub 2019 Sep 3. Glia. 2019. PMID: 31479171 Free PMC article. - Lineages to circuits: the developmental and evolutionary architecture of information channels into the central complex.
Kandimalla P, Omoto JJ, Hong EJ, Hartenstein V. Kandimalla P, et al. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2023 Jul;209(4):679-720. doi: 10.1007/s00359-023-01616-y. Epub 2023 Mar 17. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2023. PMID: 36932234 Free PMC article. Review.
References
- Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. - PubMed
- Bello BC, Hirth F, Gould AP. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–219. - PubMed
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. - PubMed
- Bier E, Vaessin H, Younger-Shepherd S, Jan LY, Jan YN. deadpan, an essential pan-neural gene in Drosophila, encodes a helix-loop-helix protein similar to the hairy gene product. Genes Dev. 1992;6:2137–2151. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous