Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant - PubMed (original) (raw)
doi: 10.1136/jmg.2007.055202. Epub 2008 Jun 11.
A J Sharp, H C Mefford, T de Ravel, C A Ruivenkamp, M H Breuning, J-P Fryns, K Devriendt, G Van Buggenhout, A Vogels, H Stewart, R C Hennekam, G M Cooper, R Regan, S J L Knight, E E Eichler, J R Vermeesch
Affiliations
- PMID: 18550696
- PMCID: PMC2658752
- DOI: 10.1136/jmg.2007.055202
Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant
F D Hannes et al. J Med Genet. 2009 Apr.
Abstract
Background: Genomic disorders are often caused by non-allelic homologous recombination between segmental duplications. Chromosome 16 is especially rich in a chromosome-specific low copy repeat, termed LCR16.
Methods and results: A bacterial artificial chromosome (BAC) array comparative genome hybridisation (CGH) screen of 1027 patients with mental retardation and/or multiple congenital anomalies (MR/MCA) was performed. The BAC array CGH screen identified five patients with deletions and five with apparently reciprocal duplications of 16p13 covering 1.65 Mb, including 15 RefSeq genes. In addition, three atypical rearrangements overlapping or flanking this region were found. Fine mapping by high-resolution oligonucleotide arrays suggests that these deletions and duplications result from non-allelic homologous recombination (NAHR) between distinct LCR16 subunits with >99% sequence identity. Deletions and duplications were either de novo or inherited from unaffected parents. To determine whether these imbalances are associated with the MR/MCA phenotype or whether they might be benign variants, a population of 2014 normal controls was screened. The absence of deletions in the control population showed that 16p13.11 deletions are significantly associated with MR/MCA (p = 0.0048). Despite phenotypic variability, common features were identified: three patients with deletions presented with MR, microcephaly and epilepsy (two of these had also short stature), and two other deletion carriers ascertained prenatally presented with cleft lip and midline defects. In contrast to its previous association with autism, the duplication seems to be a common variant in the population (5/1682, 0.29%).
Conclusion: These findings indicate that deletions inherited from clinically normal parents are likely to be causal for the patients' phenotype whereas the role of duplications (de novo or inherited) in the phenotype remains uncertain. This difference in knowledge regarding the clinical relevance of the deletion and the duplication causes a paradigm shift in (cyto)genetic counselling.
Conflict of interest statement
Competing interests: None declared.
Figures
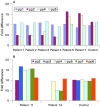
Figure 1
Molecular overview of recurrent deletions and reciprocal duplications in patients 1–10. (A) ENSEMBL overview (freeze: 24-04-2007) which visualises 1 Mb bacterial artificial chromosome (BAC) clones, the genes involved in the imbalance, the copy number variable regions and chromosome bands. (B) The location of real-time quantitative PCR primers used to fine-map the breakpoints, are depicted. (C) The extent of the deletion and duplication is shown by red and green bars, respectively in typical del/dup patients. (D) Organisation of the segmental duplication structure at the distal and proximal breakpoints of recurrent 16p13.1 rearrangements. Each coloured bar represents a pairwise alignment with >98% identity.
Figure 2
Real-time quantitative PCR results with different primer sets flanking the breakpoint regions. (A) In all individuals carrying the common deletion/duplication (patients 1–10), the distal breakpoints occur between primers pp1 and pp2, and the proximal breakpoint is defined by primers pp4 and pp5. (B) The extent of the atypical duplication and deletion identified in patients 11 and 13 were delineated by primersets pp2/pp3 and pp4/pp5, respectively, for distal breakpoints and between pp6 and pp7 for the proximal breakpoint. The presence of two copies for a locus was defined by a fold difference of 1 whereas a fold difference of 0.5 or 1.5 corresponds to a deletion or a duplication, respectively.
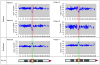
Figure 3
Detection of 16p12–p13 imbalances defined by the 262k NspI SNP array. Data from the common 16p13.11 1.65 Mb rearrangements are shown. Each plot has physical probe position on 16p (_x_-axis) against probe intensity ratio (_y_-axis). Red shading, common deleted region; green shading, duplication in patients 1–4, 6 and 7. Chr, chromosome.
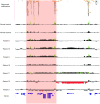
Figure 4
High-resolution oligonucleotide array mapping of seven 16p12.3–p13.11 rearrangements, of which four have a common distal breakpoint (14.7–14.75 Mb). For those four patients with the common 1.65 Mb rearrangement (red shading), the proximal breakpoints also map to a second LCR16 cluster (16.3–16.77 Mb). Another three patients have an atypical rearrangement: patients 11 and 12 show an atypical larger duplication, with the distal breakpoint between 15.0–15.4 Mb and the proximal breakpoint located within a third LCR16 cluster (18.3–18.4 Mb), and patient 13 has an atypical deletion with proximal breakpoint in the third LCR16 cluster and distal breakpoint in the second cluster. Data from normal control individuals show that there is marked copy number variation in the LCR16 clusters that define these three breakpoint regions. Note that the high degree of homology between these LCR16s also results in false-positive signals from probes that are identical to those within the true deletion/duplication in patients 5, 8 and 9. The image has a 5 Mb region of 16p12–p13 (chr16:14 000 000–19 000 000). For each individual, deviations of probe log2 ratios from zero are depicted by grey/black lines, with those exceeding a threshold of 1.5 SD from the mean probe ratio shown in green and red to represent relative gains and losses, respectively. Segmental duplications of increasing similarity (90–98%, 98–99%, and >99%) are represented by grey/yellow/orange bars, respectively.
Similar articles
- Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size.
Shinawi M, Liu P, Kang SH, Shen J, Belmont JW, Scott DA, Probst FJ, Craigen WJ, Graham BH, Pursley A, Clark G, Lee J, Proud M, Stocco A, Rodriguez DL, Kozel BA, Sparagana S, Roeder ER, McGrew SG, Kurczynski TW, Allison LJ, Amato S, Savage S, Patel A, Stankiewicz P, Beaudet AL, Cheung SW, Lupski JR. Shinawi M, et al. J Med Genet. 2010 May;47(5):332-41. doi: 10.1136/jmg.2009.073015. Epub 2009 Nov 12. J Med Genet. 2010. PMID: 19914906 Free PMC article. - Phenotypic manifestations of copy number variation in chromosome 16p13.11.
Nagamani SC, Erez A, Bader P, Lalani SR, Scott DA, Scaglia F, Plon SE, Tsai CH, Reimschisel T, Roeder E, Malphrus AD, Eng PA, Hixson PM, Kang SH, Stankiewicz P, Patel A, Cheung SW. Nagamani SC, et al. Eur J Hum Genet. 2011 Mar;19(3):280-6. doi: 10.1038/ejhg.2010.184. Epub 2010 Dec 8. Eur J Hum Genet. 2011. PMID: 21150890 Free PMC article. - Int22h-1/int22h-2-mediated Xq28 rearrangements: intellectual disability associated with duplications and in utero male lethality with deletions.
El-Hattab AW, Fang P, Jin W, Hughes JR, Gibson JB, Patel GS, Grange DK, Manwaring LP, Patel A, Stankiewicz P, Cheung SW. El-Hattab AW, et al. J Med Genet. 2011 Dec;48(12):840-50. doi: 10.1136/jmedgenet-2011-100125. Epub 2011 Oct 8. J Med Genet. 2011. PMID: 21984752 - Phenotypic expansion of the interstitial 16p13.3 duplication: a case report and review of the literature.
Li Z, Liu J, Li H, Peng Y, Lv W, Long Z, Liang D, Wu L. Li Z, et al. Gene. 2013 Dec 1;531(2):502-5. doi: 10.1016/j.gene.2013.09.006. Epub 2013 Sep 12. Gene. 2013. PMID: 24035902 Review. - 17q12 deletion and duplication syndrome in Denmark-A clinical cohort of 38 patients and review of the literature.
Rasmussen M, Vestergaard EM, Graakjaer J, Petkov Y, Bache I, Fagerberg C, Kibaek M, Svaneby D, Petersen OB, Brasch-Andersen C, Sunde L. Rasmussen M, et al. Am J Med Genet A. 2016 Nov;170(11):2934-2942. doi: 10.1002/ajmg.a.37848. Epub 2016 Jul 13. Am J Med Genet A. 2016. PMID: 27409573 Review.
Cited by
- Prenatal diagnosis and postnatal follow-up of 15 fetuses with 16p13.11 microduplication syndrome.
Zhao Y, Song L, Zhang S, Hou F, Shan S, Jin H. Zhao Y, et al. Front Genet. 2024 Oct 15;15:1486974. doi: 10.3389/fgene.2024.1486974. eCollection 2024. Front Genet. 2024. PMID: 39473442 Free PMC article. - Clinical phenotype of the 16p.13.11 microdeletion: a case report with a mini review of the literature.
Palumbi R, Ponzi E, Micella S, Pascali M, Bucci R, Gentile M, Margari L, Simone M. Palumbi R, et al. Front Genet. 2024 Aug 16;15:1429185. doi: 10.3389/fgene.2024.1429185. eCollection 2024. Front Genet. 2024. PMID: 39221225 Free PMC article. - Severe Unilateral Microtia with Aural Atresia, Hair White Patch, Stereotypes in a Young Boy with De novo 16p13.11 Deletion: Reasons for a New Genotype-Phenotype Correlation.
Pavone P, Pappalardo XG, Parano C, Parano E, Corsello A, Ruggieri M, Cacciaguerra G, Falsaperla R. Pavone P, et al. Glob Med Genet. 2023 Dec 4;10(4):370-375. doi: 10.1055/s-0043-1777362. eCollection 2023 Dec. Glob Med Genet. 2023. PMID: 38053544 Free PMC article. - Performance of noninvasive prenatal testing for twin pregnancies in South China.
Wang D, Peng H, Wang Y, Hou Y, Guo F, Zhu J, Hu T, Yang J. Wang D, et al. J Assist Reprod Genet. 2023 Sep;40(9):2219-2231. doi: 10.1007/s10815-023-02881-1. Epub 2023 Jul 22. J Assist Reprod Genet. 2023. PMID: 37480419 Free PMC article.
References
- Inoue K, Lupski JR. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet 2002;3:199–242 - PubMed
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science 2002;297:1003–7 - PubMed
- Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC, Fitzpatrick CA, Segraves R, Richmond TA, Guiver C, Albertson DG, Pinkel D, Eis PS, Schwartz S, Knight SJ, Eichler EE. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet 2006;38:1038–42 - PubMed
- Balciuniene J, Feng N, Iyadurai K, Hirsch B, Charnas L, Bill BR, Easterday MC, Staaf J, Oseth L, Czapansky-Beilman D, Avramopoulos D, Thomas GH, Borg A, Valle D, Schimmenti LA, Selleck SB. Recurrent 10q22–q23 deletions: a genomic disorder on 10q associated with cognitive and behavioral abnormalities. Am J Hum Genet 2007;80:938–47 - PMC - PubMed
- Loftus BJ, Kim UJ, Sneddon VP, Kalush F, Brandon R, Fuhrmann J, Mason T, Crosby ML, Barnstead M, Cronin L, Deslattes Mays A, Cao Y, Xu RX, Kang HL, Mitchell S, Eichler EE, Harris PC, Venter JC, Adams MD. Genome duplications and other features in 12 Mb of DNA sequence from human chromosome 16p and 16q. Genomics 1999;60:295–308 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous