On the origin of new genes in Drosophila - PubMed (original) (raw)
On the origin of new genes in Drosophila
Qi Zhou et al. Genome Res. 2008 Sep.
Abstract
Several mechanisms have been proposed to account for the origination of new genes. Despite extensive case studies, the general principles governing this fundamental process are still unclear at the whole-genome level. Here, we unveil genome-wide patterns for the mutational mechanisms leading to new genes and their subsequent lineage-specific evolution at different time nodes in the Drosophila melanogaster species subgroup. We find that (1) tandem gene duplication has generated approximately 80% of the nascent duplicates that are limited to single species (D. melanogaster or Drosophila yakuba); (2) the most abundant new genes shared by multiple species (44.1%) are dispersed duplicates, and are more likely to be retained and be functional; (3) de novo gene origination from noncoding sequences plays an unexpectedly important role during the origin of new genes, and is responsible for 11.9% of the new genes; (4) retroposition is also an important mechanism, and had generated approximately 10% of the new genes; (5) approximately 30% of the new genes in the D. melanogaster species complex recruited various genomic sequences and formed chimeric gene structures, suggesting structure innovation as an important way to help fixation of new genes; and (6) the rate of the origin of new functional genes is estimated to be five to 11 genes per million years in the D. melanogaster subgroup. Finally, we survey gene frequencies among 19 globally derived strains for D. melanogaster-specific new genes and reveal that 44.4% of them show copy number polymorphisms within a population. In conclusion, we provide a panoramic picture for the origin of new genes in Drosophila species.
Figures
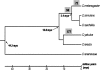
Figure 1.
Phylogenetic distribution of new genes. We designated numbers of new genes restricted to different lineages. Phylogenetic tree of investigated Drosophila species and their divergence times in each node are indicated (Tamura et al. 2004).
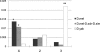
Figure 2.
Quantification of contributions to new gene origination from different mechanisms. (D. mel) D. melanogaster, (D. sch) D. sechellia, (D. sim) D. simulans. These three species together stand for the D. melanogaster species complex. We didn’t identify new genes originating from retroposition or de novo processes in the D. yakuba (D. yak) lineage due to the lack of reliable annotations in this species.
Figure 3.
Decrease of proportion of complete duplicated new genes with time. (D. mel) D. melanogaster, (D. sch) D. sechellia, (D. sim) D. simulans. These three species together stand for the D. melanogaster species complex. We compared structures and lengths of parental and new genes in D. melanogaster and the D. melanogaster species complex. New genes completely duplicating their ancestors’ coding regions seem more vulnerable to subsequent loss or structure changes during evolution.
Figure 4.
Chromosomal distribution of new genes in three datasets. New gene numbers were divided by total gene numbers on the specific chromosome as a normalization. We marked significant (*P < 0.01, Fisher’s exact test) and highly significant (**P < 0.001) overrepresentation with of new genes on certain chromosomes. We didn’t take chromosome 4 into account given the extremely low gene number on this chromosome.
Similar articles
- Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila.
Bai Y, Casola C, Feschotte C, Betrán E. Bai Y, et al. Genome Biol. 2007;8(1):R11. doi: 10.1186/gb-2007-8-1-r11. Genome Biol. 2007. PMID: 17233920 Free PMC article. - Duplication-degeneration as a mechanism of gene fission and the origin of new genes in Drosophila species.
Wang W, Yu H, Long M. Wang W, et al. Nat Genet. 2004 May;36(5):523-7. doi: 10.1038/ng1338. Epub 2004 Apr 4. Nat Genet. 2004. PMID: 15064762 - Formation and longevity of chimeric and duplicate genes in Drosophila melanogaster.
Rogers RL, Bedford T, Hartl DL. Rogers RL, et al. Genetics. 2009 Jan;181(1):313-22. doi: 10.1534/genetics.108.091538. Epub 2008 Nov 17. Genetics. 2009. PMID: 19015547 Free PMC article. - Patterns of polymorphism and divergence from noncoding sequences of Drosophila melanogaster and D. simulans: evidence for nonequilibrium processes.
Kern AD, Begun DJ. Kern AD, et al. Mol Biol Evol. 2005 Jan;22(1):51-62. doi: 10.1093/molbev/msh269. Epub 2004 Sep 29. Mol Biol Evol. 2005. PMID: 15456897 Review. - New gene evolution: little did we know.
Long M, VanKuren NW, Chen S, Vibranovski MD. Long M, et al. Annu Rev Genet. 2013;47:307-33. doi: 10.1146/annurev-genet-111212-133301. Epub 2013 Sep 13. Annu Rev Genet. 2013. PMID: 24050177 Free PMC article. Review.
Cited by
- The Recent De Novo Origin of Protein C-Termini.
Andreatta ME, Levine JA, Foy SG, Guzman LD, Kosinski LJ, Cordes MH, Masel J. Andreatta ME, et al. Genome Biol Evol. 2015 May 21;7(6):1686-701. doi: 10.1093/gbe/evv098. Genome Biol Evol. 2015. PMID: 26002864 Free PMC article. - De novo activated transcription of inserted foreign coding sequences is inheritable in the plant genome.
Hata T, Takada N, Hayakawa C, Kazama M, Uchikoba T, Tachikawa M, Matsuo M, Satoh S, Obokata J. Hata T, et al. PLoS One. 2021 Jun 10;16(6):e0252674. doi: 10.1371/journal.pone.0252674. eCollection 2021. PLoS One. 2021. PMID: 34111139 Free PMC article. - In with the old, in with the new: the promiscuity of the duplication process engenders diverse pathways for novel gene creation.
Katju V. Katju V. Int J Evol Biol. 2012;2012:341932. doi: 10.1155/2012/341932. Epub 2012 Sep 13. Int J Evol Biol. 2012. PMID: 23008799 Free PMC article. - Preferential duplication of intermodular hub genes: an evolutionary signature in eukaryotes genome networks.
Ferreira RM, Rybarczyk-Filho JL, Dalmolin RJ, Castro MA, Moreira JC, Brunnet LG, de Almeida RM. Ferreira RM, et al. PLoS One. 2013;8(2):e56579. doi: 10.1371/journal.pone.0056579. Epub 2013 Feb 26. PLoS One. 2013. PMID: 23468868 Free PMC article. - The HIV-1 Antisense Gene ASP: The New Kid on the Block.
Gholizadeh Z, Iqbal MS, Li R, Romerio F. Gholizadeh Z, et al. Vaccines (Basel). 2021 May 17;9(5):513. doi: 10.3390/vaccines9050513. Vaccines (Basel). 2021. PMID: 34067514 Free PMC article. Review.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
- Arguello J.R., Chen Y., Yang S., Wang W., Long M., Chen Y., Yang S., Wang W., Long M., Yang S., Wang W., Long M., Wang W., Long M., Long M. Origination of an X-linked testes chimeric gene by illegitimate recombination in Drosophila. PLoS Genet. 2006;2:e77. doi: 10.1371/journal.pgen.0020077. - DOI - PMC - PubMed
- Bai Y., Casola C., Feschotte C., Betran E., Casola C., Feschotte C., Betran E., Feschotte C., Betran E., Betran E. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 2007;8:R11. doi: 10.1186/gb-2007-8-1-r11. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases