Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana - PubMed (original) (raw)
Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana
Sascha Laubinger et al. Proc Natl Acad Sci U S A. 2008.
Abstract
The processing of Arabidopsis thaliana microRNAs (miRNAs) from longer primary transcripts (pri-miRNAs) requires the activity of several proteins, including DICER-LIKE1 (DCL1), the double-stranded RNA-binding protein HYPONASTIC LEAVES1 (HYL1), and the zinc finger protein SERRATE (SE). It has been noted before that the morphological appearance of weak se mutants is reminiscent of plants with mutations in ABH1/CBP80 and CBP20, which encode the two subunits of the nuclear cap-binding complex. We report that, like SE, the cap-binding complex is necessary for proper processing of pri-miRNAs. Inactivation of either ABH1/CBP80 or CBP20 results in decreased levels of mature miRNAs accompanied by apparent stabilization of pri-miRNAs. Whole-genome tiling array analyses reveal that se, abh1/cbp80, and cbp20 mutants also share similar splicing defects, leading to the accumulation of many partially spliced transcripts. This is unlikely to be an indirect consequence of improper miRNA processing or other mRNA turnover pathways, because introns retained in se, abh1/cbp80, and cbp20 mutants are not affected by mutations in other genes required for miRNA processing or for nonsense-mediated mRNA decay. Taken together, our results uncover dual roles in splicing and miRNA processing that distinguish SE and the cap-binding complex from specialized miRNA processing factors such as DCL1 and HYL1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
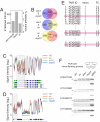
Requirement of the CBC for pri-miRNA processing. (A) Comparative gross morphology of 21-day-old wild-type (WT), _abh1_-285, _abh1_-753, and _se_-1 plants. (B) Detection of selected miRNAs by RNA blot in 10-day-old wild-type, _abh1_-285, _abh1_-753, and _se_-1 plants. Ethidium-stained gels on which tRNA and rRNA bands are visible are shown as loading control. (C) Comparative gross morphology of 21-day-old wild-type, _se_-1, _abh1_-285, and cbp20 plants. (D) Detection of selected miRNAs in 10-day-old wild-type, _se_-1, _abh1_-285, and cbp20 plants. (E) Real-time RT-PCR analysis of selected pri-miRNAs in wild-type, _se_-1, _se_-3, _abh1_-285, _abh1_-753, cbp20, and hyl1 plants. (F) Overlap of pri-miRNAs that accumulate to higher levels in abh1/_cbp80_-285, cbp20 and _se_-1 mutants. (G) As two examples for pri-miRNAs that accumulate in mutants, hybridization intensities on tiling arrays are shown for pri-miR160c and pri-miR172a. Values are averaged from three biological replicates each. Accumulation in _se_-1 is apparent for both pri-miRNA transcripts, but only pri-miR160c appears to be affected also by abh1 and cbp20 mutations. Tick marks indicate 100 bases each.
Fig. 2.
Splicing defects in abh1/_cbp80_-285, cbp20 and _se_-1 mutants. (A) Number of retained introns in mutants. (B) Overlap of retained introns among the three mutants examined. Parentheses give overlap expected by chance. (C) As one example for an improperly spliced mRNA detected by tiling arrays, hybridization signals of probes representing the AT3G01500 gene are shown for wild-type, abh1/cbp80, cbp20 and se plants. Values are averaged from three biological replicates each. Gray box highlights retained intron. Annotated splice forms (TAIR 7) are shown below, with blue indicating coding sequence. Tick marks indicate 500 bases each. (D) As a second example, tiling array signals for AT3G04670 are shown. Note second intron (purple underline) that is unchanged in the mutants. (E) Excerpt from table of genes with introns retained in abh1/cbp80, cbp20, and _se_-1 mutants (see
Table S2
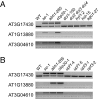
). The second column (“Intron”) indicates intron position from 5′ end. As discussed in the text, first introns are overrepresented. FC, fold change. Details for highlighted genes are shown in F. (F) Validation of intron retention in selected genes by conventional RT-PCR analysis. gDNA, genomic DNA control. “+” and “−” indicates reactions with and without reverse transcriptase. The upper band corresponds to unspliced form (as in gDNA), the lower band to spliced form.
Fig. 3.
Analysis of splicing in other miRNA-processing mutants and in NMD mutants. (A) Comparative RT-PCR analysis of mRNAs with introns retained in abh1/cbp80, cbp20 and se mutants in other miRNA processing mutants and related genotypes (dcl1, dcl2 dcl3 dcl4, hyl1, hst and ago1). (B) Comparative RT-PCR analysis of NMD mutants (upf1 and upf3).
Fig. 4.
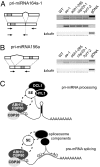
Accumulation of intron-containing pri-miRNAs. (A) Diagram of the pri-miRNA164a-1 transcript (53), with miRNA foldback indicated in gray, with intron-flanking and exon–exon-junction spanning oligonucleotide primers shown as arrows. RT-PCR results are shown on the right. (B) Diagram of the pri-miRNA156a transcript (52) and RT-PCR analysis. (C) Illustration comparing potential roles of CBC and SE in pri-miRNA processing and pre-mRNA splicing. SE might be part of a larger cap-binding complex (indicated by an additional component with a dashed outline) or might interact only transiently with the CPC.
Comment in
- Splicing and dicing with a SERRATEd edge.
Montgomery TA, Carrington JC. Montgomery TA, et al. Proc Natl Acad Sci U S A. 2008 Jun 24;105(25):8489-90. doi: 10.1073/pnas.0804356105. Epub 2008 Jun 16. Proc Natl Acad Sci U S A. 2008. PMID: 18559848 Free PMC article. No abstract available.
Similar articles
- Dissecting the interactions of SERRATE with RNA and DICER-LIKE 1 in Arabidopsis microRNA precursor processing.
Iwata Y, Takahashi M, Fedoroff NV, Hamdan SM. Iwata Y, et al. Nucleic Acids Res. 2013 Oct;41(19):9129-40. doi: 10.1093/nar/gkt667. Epub 2013 Aug 5. Nucleic Acids Res. 2013. PMID: 23921632 Free PMC article. - SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis.
Yang L, Liu Z, Lu F, Dong A, Huang H. Yang L, et al. Plant J. 2006 Sep;47(6):841-50. doi: 10.1111/j.1365-313X.2006.02835.x. Epub 2006 Aug 2. Plant J. 2006. PMID: 16889646 - Splicing and dicing with a SERRATEd edge.
Montgomery TA, Carrington JC. Montgomery TA, et al. Proc Natl Acad Sci U S A. 2008 Jun 24;105(25):8489-90. doi: 10.1073/pnas.0804356105. Epub 2008 Jun 16. Proc Natl Acad Sci U S A. 2008. PMID: 18559848 Free PMC article. No abstract available. - mRNA cap binding proteins: effects on abscisic acid signal transduction, mRNA processing, and microarray analyses.
Kuhn JM, Hugouvieux V, Schroeder JI. Kuhn JM, et al. Curr Top Microbiol Immunol. 2008;326:139-50. doi: 10.1007/978-3-540-76776-3_8. Curr Top Microbiol Immunol. 2008. PMID: 18630751 Review. - New insights into pri-miRNA processing and accumulation in plants.
Zhang S, Liu Y, Yu B. Zhang S, et al. Wiley Interdiscip Rev RNA. 2015 Sep-Oct;6(5):533-45. doi: 10.1002/wrna.1292. Epub 2015 Jun 29. Wiley Interdiscip Rev RNA. 2015. PMID: 26119101 Review.
Cited by
- A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis.
Wu X, Shi Y, Li J, Xu L, Fang Y, Li X, Qi Y. Wu X, et al. Cell Res. 2013 May;23(5):645-57. doi: 10.1038/cr.2013.23. Epub 2013 Feb 12. Cell Res. 2013. PMID: 23399598 Free PMC article. - SERRATE is required for intron suppression of RNA silencing in Arabidopsis.
Christie M, Carroll BJ. Christie M, et al. Plant Signal Behav. 2011 Dec;6(12):2035-7. doi: 10.4161/psb.6.12.18238. Plant Signal Behav. 2011. PMID: 22112452 Free PMC article. - Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses.
Li W, Cui X, Meng Z, Huang X, Xie Q, Wu H, Jin H, Zhang D, Liang W. Li W, et al. Plant Physiol. 2012 Mar;158(3):1279-92. doi: 10.1104/pp.111.188789. Epub 2012 Jan 13. Plant Physiol. 2012. PMID: 22247272 Free PMC article. - MIR846 and MIR842 comprise a cistronic MIRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis.
Jia F, Rock CD. Jia F, et al. Plant Mol Biol. 2013 Mar;81(4-5):447-60. doi: 10.1007/s11103-013-0015-6. Epub 2013 Jan 23. Plant Mol Biol. 2013. PMID: 23341152 Free PMC article. - Expression of microRNAs and its regulation in plants.
Xie Z, Khanna K, Ruan S. Xie Z, et al. Semin Cell Dev Biol. 2010 Oct;21(8):790-7. doi: 10.1016/j.semcdb.2010.03.012. Epub 2010 Apr 18. Semin Cell Dev Biol. 2010. PMID: 20403450 Free PMC article. Review.
References
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. - PubMed
- Finnegan EJ, Margis R, Waterhouse PM. Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr Biol. 2003;13:236–240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous