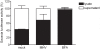Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation - PubMed (original) (raw)
Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation
Monique H Verheije et al. PLoS Pathog. 2008.
Abstract
Coronaviruses induce in infected cells the formation of double membrane vesicles, which are the sites of RNA replication. Not much is known about the formation of these vesicles, although recent observations indicate an important role for the endoplasmic reticulum in the formation of the mouse hepatitis coronavirus (MHV) replication complexes (RCs). We now show that MHV replication is sensitive to brefeldin A (BFA). Consistently, expression of a dominant-negative mutant of ARF1, known to mimic the action of the drug, inhibited MHV infection profoundly. Immunofluorescence analysis and quantitative electron microscopy demonstrated that BFA did not block the formation of RCs per se, but rather reduced their number. MHV RNA replication was not sensitive to BFA in MDCK cells, which are known to express the BFA-resistant guanine nucleotide exchange factor GBF1. Accordingly, individual knockdown of the Golgi-resident targets of BFA by transfection of small interfering RNAs (siRNAs) showed that GBF1, but not BIG1 or BIG2, was critically involved in MHV RNA replication. ARF1, the cellular effector of GBF1, also appeared to be involved in MHV replication, as siRNAs targeting this small GTPase inhibited MHV infection significantly. Collectively, our results demonstrate that GBF1-mediated ARF1 activation is required for efficient MHV RNA replication and reveal that the early secretory pathway and MHV replication complex formation are closely connected.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
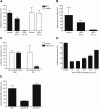
Figure 1. BFA inhibits MHV replication in mouse LR7 cells.
(A, B, D) LR7 cells were inoculated with MHV-EFLM or with Sindbis pseudovirus particles containing a luciferase replicon and incubated with 5 µg/ml BFA during the indicated time periods. At the end of each incubation period, virus replication was analyzed by determining the luciferase expression level (A and D) or the amount of viral genomic RNA (B) as described in the Material and Methods. (C) LR7 cells were inoculated with MHV-2aFLS, transfected with synthetic RNA transcribed from pM5f-RL-M3, and incubated from 2–6 h p.i. in the presence or absence of 5 µg/ml BFA. Renilla (RL) and firefly (FL) luciferase expression levels were determined in the cell lysates at 6 h p.i. and are depicted relative to untreated samples; (E) LR7 cells were transfected with pARF1-YFP, pARF1T31N-YFP, or pARF1Q71L-YFP and inoculated with MHV-RFP (moi of 1) 24 h later. At 18 h p.i. FACS analyses were performed as described in Materials and Methods. The percentages of GFP/YFP positive cells that were also RFP positive were determined relative to wild type ARF1 expressing cells. The results of representative experiments performed in triplicate are shown. Error bars indicate standard deviations.
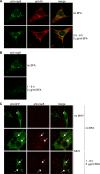
Figure 2. Immunofluorescence analysis of MHV RCs.
LR7 cells were inoculated with MHV-A59 and subsequently mock-treated (panel A, upper row), treated with 5 µg/ml BFA from 5.5–6 h p.i. (panel A, lower row) or from 1–6 h p.i. (panel B). Immunostaining was performed using antibodies against nsp8 (anti-nsp8) and against the M protein (anti-M). LR7 cells were transfected with pN-EGFP and subsequently mock-infected (panel C, upper row), infected with MHV-A59 (panel C, middle row), or infected with MHV-A59 and treated with 5 µg/ml BFA from 1 to 7 h p.i. (panel C, bottom row). At 7 h p.i., cells were fixed and an immunostaining was performed using the nsp8 antibodies. Identical confocal microscopy settings were used for mock-treated and BFA-treated samples. Arrowheads in panel C indicate cytosolic staining; arrows indicate nsp8-positive foci.
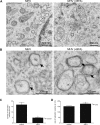
Figure 3. Ultrastructural analysis of MHV-infected LR7 cells.
LR7 cells were inoculated with MHV-A59 and treated with or without 5 µg/ml BFA from 1–6 h p.i, chemically fixed and embedded with Epon resin. (A) Numerous clusters of virus-induced DMVs (indicated by *) were found in the perinuclear region of the cell (N-nucleus; M-mitochondrion); Panel B shows a close view of DMVs, clearly demonstrating the presence of double membranes (indicated by arrows); (C) The average number of DMVs per cell obtained by counting 20 infected cells; (D) Average DMV diameter obtained measuring 38 of them. Error bars indicate standard error of the mean (SEM).
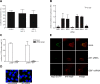
Figure 4. The role of Golgi-residing GEFs in MHV replication.
(A) MDCK(MHVR) cells were inoculated with MHV-EFLM and incubated with 5 µg/ml BFA during the indicated time periods. At 7 h p.i. the luciferase expression levels were determined; (B) HeLa-CEACAM1a cells were transfected with three siRNAs directed against either GBF1, BIG1, BIG2, ARF1, firefly luciferase (luc), or GAPDH or were mock transfected (mock). Seventy-two h post transfection, the cells were inoculated with MHV-2aFLS. At 6 h p.i., the cell viability and luciferase expression levels were measured as described in the Materials and Methods. The graph depicts the relative luciferase expression (RII) compared to mock-treated cells after correction for cell viability; (C) HeLa-CEACAM1a cells were transfected with plasmids pGBF1-YFP and pARF1-YFP in the presence or absence of their corresponding siRNAs. At 24 h post transfection, the cells were fixed and the percentage of YFP-positive cells was determined; (D) HeLa-CEACAM1a cells transfected with siRNAs targeting ARF1 and mock-transfected cells were fixed at 72 h post transfection and processed for immunostaining using antibodies against the Golgi marker GM130. (E) HeLa-CEACAM1a cells were transfected with siRNAs directed against GBF1 or ARF1, or were mock transfected. Seventy-two h post transfection, the cells were inoculated with MHV-nsp2GFP and at 6 h p.i. they were fixed and processed for immunofluorescence using the nsp8 antibody. (A–C) The results of a representative experiment performed in triplicate are shown. Error bars indicate standard deviations. (D–E) Representative images are shown.
Figure 5. ARF1 and COPI do not co-localize with the RCs.
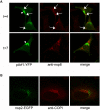
(A) LR7 cells were transfected with pARF1-YFP, pARF1T31N-YFP, or pARF1Q71L-YFP and inoculated with MHV-A59 (moi of 1) 24 h later. At 4 h and 7 h p.i. cells were processed for immunofluorescence using antibodies against nsp8. Arrows indicate co-localization of nsp8 with ARF1; arrowheads indicate ARF1 localizing to the Golgi complex; (B) HeLa-CEACAM1a cells were inoculated with MHV-nsp2-GFP (moi of 1), fixed 7 h later and processed for immunofluorescence using antibodies against αγCOPI.
Figure 6. MHV reduces but does not block protein secretion.
LR7 cells, transfected with a plasmid encoding the Gaussia gene, were at 1 h post transfection either infected with MHV-A59 or mock-infected or were treated with BFA. At 4.5 h p.i. Gaussia luciferase activity was determined both in the cell lysate and in the culture supernatant. The relative amount of luciferase present in the supernatant and the cell lysate is depicted.
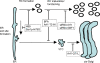
Figure 7. Model of MHV RCs and their links to the early secretory pathway.
Two major steps in the anterograde protein secretion route (reviewed in [81]) are linked to MHV RC formation and/or RNA replication. First, transport of proteins out of the ER requires ER exit site formation controlled by Sar1p ,,. Blocking this early step by using the drug H89 or by expressing of a dominant mutant of Sar1p blocks MHV replication profoundly . Next, ER exit sites develop into, or form de novo, vesicular-tubular clusters (VTCs) (also called ERGIC), for which GBF1 and ARF1 are required. This step, which can be blocked by BFA, by expressing a dominant-negative mutant of ARF1 or by down-regulating ARF1 using siRNAs , is also involved in MHV RC formation (this manuscript). However, a fully functional secretory pathway is not essential, as a dominant-active mutant of ARF1, which blocks transport between VTCs and cis-Golgi , does not impair MHV replication.
Similar articles
- GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication.
Lanke KH, van der Schaar HM, Belov GA, Feng Q, Duijsings D, Jackson CL, Ehrenfeld E, van Kuppeveld FJ. Lanke KH, et al. J Virol. 2009 Nov;83(22):11940-9. doi: 10.1128/JVI.01244-09. Epub 2009 Sep 9. J Virol. 2009. PMID: 19740986 Free PMC article. - Rab1b-GBF1-ARF1 Secretory Pathway Axis Is Required for Birnavirus Replication.
Gimenez MC, Frontini-Lopez YR, Pocognoni CA, Roldán JS, García Samartino C, Uhart M, Colombo MI, Terebiznik MR, Delgui LR. Gimenez MC, et al. J Virol. 2022 Feb 23;96(4):e0200521. doi: 10.1128/JVI.02005-21. Epub 2021 Dec 8. J Virol. 2022. PMID: 34878889 Free PMC article. - Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication.
Goueslain L, Alsaleh K, Horellou P, Roingeard P, Descamps V, Duverlie G, Ciczora Y, Wychowski C, Dubuisson J, Rouillé Y. Goueslain L, et al. J Virol. 2010 Jan;84(2):773-87. doi: 10.1128/JVI.01190-09. Epub 2009 Nov 11. J Virol. 2010. PMID: 19906930 Free PMC article. - Role of the Guanine Nucleotide Exchange Factor GBF1 in the Replication of RNA Viruses.
Martínez JL, Arias CF. Martínez JL, et al. Viruses. 2020 Jun 24;12(6):682. doi: 10.3390/v12060682. Viruses. 2020. PMID: 32599855 Free PMC article. Review. - Coronavirus infection of polarized epithelial cells.
Rossen JW, Horzinek MC, Rottier PJ. Rossen JW, et al. Trends Microbiol. 1995 Dec;3(12):486-90. doi: 10.1016/s0966-842x(00)89018-6. Trends Microbiol. 1995. PMID: 8800844 Free PMC article. Review.
Cited by
- Quantitative Proteomics of Uukuniemi Virus-host Cell Interactions Reveals GBF1 as Proviral Host Factor for Phleboviruses.
Uckeley ZM, Moeller R, Kühn LI, Nilsson E, Robens C, Lasswitz L, Lindqvist R, Lenman A, Passos V, Voss Y, Sommerauer C, Kampmann M, Goffinet C, Meissner F, Överby AK, Lozach PY, Gerold G. Uckeley ZM, et al. Mol Cell Proteomics. 2019 Dec;18(12):2401-2417. doi: 10.1074/mcp.RA119.001631. Epub 2019 Sep 30. Mol Cell Proteomics. 2019. PMID: 31570497 Free PMC article. - SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum.
Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. Knoops K, et al. PLoS Biol. 2008 Sep 16;6(9):e226. doi: 10.1371/journal.pbio.0060226. PLoS Biol. 2008. PMID: 18798692 Free PMC article. - Development and RNA-synthesizing activity of coronavirus replication structures in the absence of protein synthesis.
van den Worm SH, Knoops K, Zevenhoven-Dobbe JC, Beugeling C, van der Meer Y, Mommaas AM, Snijder EJ. van den Worm SH, et al. J Virol. 2011 Jun;85(11):5669-73. doi: 10.1128/JVI.00403-11. Epub 2011 Mar 23. J Virol. 2011. PMID: 21430047 Free PMC article. - Identification of host factors involved in coronavirus replication by quantitative proteomics analysis.
Vogels MW, van Balkom BW, Kaloyanova DV, Batenburg JJ, Heck AJ, Helms JB, Rottier PJ, de Haan CA. Vogels MW, et al. Proteomics. 2011 Jan;11(1):64-80. doi: 10.1002/pmic.201000309. Epub 2010 Dec 6. Proteomics. 2011. PMID: 21182195 Free PMC article. - On the Host Side of the Hepatitis E Virus Life Cycle.
Oechslin N, Moradpour D, Gouttenoire J. Oechslin N, et al. Cells. 2020 May 22;9(5):1294. doi: 10.3390/cells9051294. Cells. 2020. PMID: 32456000 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources