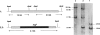Transfer of the first arabinofuranose residue to galactan is essential for Mycobacterium smegmatis viability - PubMed (original) (raw)
Transfer of the first arabinofuranose residue to galactan is essential for Mycobacterium smegmatis viability
Libin Shi et al. J Bacteriol. 2008 Aug.
Abstract
The mycobacterial arabinan is an elaborate component of the cell wall with multiple glycosyl linkages and no repeating units. In Mycobacterium spp., the Emb proteins (EmbA, EmbB, and EmbC) have been identified as putative mycobacterial arabinosyltransferases implicated in the biogenesis of the cell wall arabinan. Furthermore, it is now evident that the EmbA and EmbB proteins are involved in the assembly of the nonreducing terminal motif of arabinogalactan and EmbC is involved in transferring arabinose, perhaps in the early stage of arabinan synthesis in lipoarabinomannan. It has also been shown that the Emb proteins are a target of the antimycobacterial drug ethambutol (EMB). In the search for additional mycobacterial arabinosyltransferases in addition to the Emb proteins, we disrupted MSMEG_6386 (an orthologue of Rv3792 and a gene upstream of embC) in Mycobacterium smegmatis. Allelic exchange at the chromosomal MSMEG_6386 locus of M. smegmatis could only be achieved in the presence of a rescue plasmid carrying a functional copy of MSMEG_6386 or Rv3792, strongly suggesting that MSMEG_6386 is essential. An in vitro arabinosyltransferase assay using a membrane preparation from M. smegmatis expressing Rv3792 and synthetic beta-d-Galf-(1-->5)-beta-D-Galf-(1-->6)-beta-D-Galf-octyl and beta-D-Galf-(1-->6)-beta-D-Galf-(1-->5)-beta-D-Galf-octyl showed that Rv3792 gene product can transfer an arabinose residue to the C-5 position of the internal 6-linked galactose. The reactions were insensitive to EMB, and when alpha-d-Manp-(1-->6)-alpha-D-Manp-(1-->6)-alpha-D-Manp-octylthiomethyl was used as an acceptor, no product was formed. These observations indicate that transfer of the first arabinofuranose residue to galactan is essential for M. smegmatis viability.
Figures
FIG. 1.
Only the nonreducing end arabinan domains in LAM and AG are shown here. Arabinan consists of the Ara2 internal motifs and Ara4 and Ara6 terminal motifs. The Ara2 motif consists of α-
d
-Ara_f_-(1→5)-α-
d
-Ara_f_ and forms the linear backbone of arabinan, the Ara6 motif is [β-
d
-Ara_f_-(1→2)-α-
d
-Ara_f_-(1→)]2-(3,5)-α-
d
-Ara_f_-(1→5)-α-
d
-Ara_f_ (A), and the Ara4 motif is β-
d
-Ara_f_-(1→2)-α-
d
-Ara_f_-(1→5)-α-
d
-Ara_f_-(1→5)-α-
d
-Ara_f_ (B). The Ara6 motif is the most characteristic structural feature in AG: both terminal β-
d
-Ara_f_ and penultimate 2-α-Ara_f_ can be covalently linked to mycolic acids, while Ara4 motifs are not found in AG. On the other hand, both Ara4 and Ara6 motifs can be found in LAM, and with various degrees of mannose capping at the nonreducing Ara4 and Ara6 termini, the arabinan in LAM plays a pivotal role in the pathogenesis of the disease.
FIG. 2.
Allelic exchange at the MSMEG_6386 locus. Southern blot analysis and expected hybridization profiles of M. smegmatis chromosomal DNA (lane 1) and the conditional mutant M. smegmatis LL2 and LL3 chromosomal DNA (lanes 2 and 3). ApaI was used to digest the chromosomal DNA. The probe used corresponds to the 2.1-kb fragment of PCRI -II generated with the primers used in the initial step. The signal detected corresponds to the 2.0-kb and 9.0-kb fragments (lane 2) and the 12.0-kb fragment (lane 3), which were carried by the rescue plasmid.
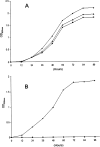
FIG. 3.
Growth characteristics of the M. smegmatis wild type and the MSMEG_6386 conditional mutants at 30°C (A) and 42°C (B). Shown are growth curves for wild-type M. smegmatis (⧫), M. smegmatis LL2 (▪), and M. smegmatis LL3 (▴) cultivated in LB-Tween 80 and LB-Tween 80-Hyg broth, respectively.
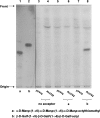
FIG. 4.
Effect of overexpressed Rv3792 in M. smegmatis on the in vitro incorporation of [14C]Ara_f_ into the synthetic trigalactan or trimannan acceptors. In lanes 1 and 2, the acceptors α-
d
-Man_p_-(1→6)-α-
d
-Man_p_-(1→6)-α-
d
-Man_p_-(CH2)6SMe (lane 1) and β-
d
-Gal-(1→5)-β-
d
-Gal_f_-(1→6)-β-
d
-Gal_f_-octyl (lane 2) were visualized by α-naphthol-sulfuric acid. Lanes 3 and 4 show the control reaction (no acceptor) from M. smegmatis with pVV16 (lane 3) and overexpressed Rv3792 (lane 4). Lanes 5 and 6 show use of α-
d
-Man_p_-(1→6)-α-
d
-Man_p_-(1→6)-α-
d
-Man_p_-(CH2)6SMe and M. smegmatis with pVV16 (lane 5) and overexpressed Rv3792 (lane 6). Lanes 7 and 8 show the incorporation of [14C]Ara_f_ into β-
d
-Gal_f_-(1→5)-β-
d
-Gal_f_-(1→6)-β-
d
-Gal_f_-octyl from M. smegmatis with pVV16 (lane 7) and overexpressed Rv3792 (lane 8). The partially purified labeled products were applied to TLC plates and developed in CHCl3-CH3OH-1 M NH4OAc-NH4OH-H2O (180:140:9:9:23 [vol/vol/vol/vol/vol]) and subjected to autoradiography.
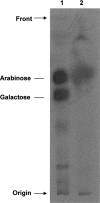
FIG. 5.
Monosaccharide analysis of the radiolabeled product after TFA hydrolysis. Radioactive AG (2,000 dpm; lane 1) and the radiolabeled product from the reaction of the incorporation of [14C]Ara_f_ into β-
d
-Gal_f_-(1→5)-β-
d
-Gal_f_-(1→6)-β-
d
-Gal_f_-octyl (lane 2) from M. smegmatis with overexpressed Rv3792 were hydrolyzed with 2 M TFA, applied to the TLC plate, developed in pyridine-ethyl acetate-acetic acid-water (5:5:1:3 [vol/vol/vol/vol]), and subjected to autoradiography.
FIG. 6.
MALDI-TOF analysis of enzymatic product. Product formed with β-
d
-Gal_f_-(1→5)-β-
d
-Gal_f_-(1→6)-β-
d
-Gal_f_-octyl extracted from a preparative TLC was methylated, and MALDI-TOF analysis of this revealed a strong molecular ion at m/z 939.5 (M + Na+) for Ara_f_-(1-?)- added to the acceptors. Intens., intensity; a.u., arbitrary units.
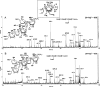
FIG. 7.
MALDI-TOF/TOF MS/MS analysis. Shown are the enzymatic product Gal3Ara1-C8H17 obtained from using the synthetic acceptor β-
d
-Gal_f_-(1→5)-β-
d
-Gal_f_-(1→6)-β-
d
-Gal_f_-octyl (A) and β-
d
-Gal_f_-(1→6)-β-
d
-Gal_f_-(1→5)-β-
d
-Gal_f_-octyl (B). The [M + Na]+ molecular ions afforded by the permethylated sample at m/z 939 (Fig. 6) were selected for high-energy CID MS/MS. Assignment of the key fragment ions were as schematically illustrated. In addition to the well-established A, X, C, and Y ions, the G and E ions were named by Spina et al. and have been described in full in the context of furanoses (18, 32). (Note that the 1,4X ions give linkage information only pertaining to the C-1 position. The 0,2X ions provide information about whether C-2 contains a glycosidic bond. 0,3A and 2,4A ions provide the C-5 and C-3 linkage information. C and Y ions, as well as the E and G ions, represent concerted elimination of substituents around the ring.) The two specific ion series corresponding to E − 30 mass units and C − 16 mass units (m/z 447 and 607, respectively) were likewise identified previously for the arabinan and not further drawn out here. The concerted cleavages corresponding to loss of both C-5 and C-6 substituents on the Gal_f_ have not been reported before and were noted to produce a pair of ions differing in having either unsaturated or saturated bonds (e.g., m/z 675/673 in panel A and 513/515 in panel B). The 1,4X ions at m/z 385 in panel A and 545 in panel B coincide, respectively, with the E ions at m/z 415 and 575 − 30 mass units and thus may not be taken as sequence-specific ions to indicate presence of isomeric products. In contrast, other specific ions as described in the text are present only in either and not both spectra. The ion at m/z 505 in panel A may be assigned as an O,3A ion, which is indicative of possible presence of Ara_f_ on the nonreducing terminal Gal2, as in panel B, but other supporting ions that are specific to this isomeric structure are lacking.
Similar articles
- The role of the embA and embB gene products in the biosynthesis of the terminal hexaarabinofuranosyl motif of Mycobacterium smegmatis arabinogalactan.
Escuyer VE, Lety MA, Torrelles JB, Khoo KH, Tang JB, Rithner CD, Frehel C, McNeil MR, Brennan PJ, Chatterjee D. Escuyer VE, et al. J Biol Chem. 2001 Dec 28;276(52):48854-62. doi: 10.1074/jbc.M102272200. Epub 2001 Oct 24. J Biol Chem. 2001. PMID: 11677227 - Characterization of a distinct arabinofuranosyltransferase in Mycobacterium smegmatis.
Zhang J, Khoo KH, Wu SW, Chatterjee D. Zhang J, et al. J Am Chem Soc. 2007 Aug 8;129(31):9650-62. doi: 10.1021/ja070330k. Epub 2007 Jul 14. J Am Chem Soc. 2007. PMID: 17630736 - The arabinosyltransferase EmbC is inhibited by ethambutol in Mycobacterium tuberculosis.
Goude R, Amin AG, Chatterjee D, Parish T. Goude R, et al. Antimicrob Agents Chemother. 2009 Oct;53(10):4138-46. doi: 10.1128/AAC.00162-09. Epub 2009 Jul 13. Antimicrob Agents Chemother. 2009. PMID: 19596878 Free PMC article. - Biosynthesis of D-arabinose in mycobacteria - a novel bacterial pathway with implications for antimycobacterial therapy.
Wolucka BA. Wolucka BA. FEBS J. 2008 Jun;275(11):2691-711. doi: 10.1111/j.1742-4658.2008.06395.x. Epub 2008 Apr 15. FEBS J. 2008. PMID: 18422659 Review. - Cell wall synthesizing complexes in Mycobacteriales.
Meyer FM, Bramkamp M. Meyer FM, et al. Curr Opin Microbiol. 2024 Jun;79:102478. doi: 10.1016/j.mib.2024.102478. Epub 2024 Apr 22. Curr Opin Microbiol. 2024. PMID: 38653035 Review.
Cited by
- The cell envelope glycoconjugates of Mycobacterium tuberculosis.
Angala SK, Belardinelli JM, Huc-Claustre E, Wheat WH, Jackson M. Angala SK, et al. Crit Rev Biochem Mol Biol. 2014 Sep-Oct;49(5):361-99. doi: 10.3109/10409238.2014.925420. Epub 2014 Jun 10. Crit Rev Biochem Mol Biol. 2014. PMID: 24915502 Free PMC article. Review. - Structure of the priming arabinosyltransferase AftA required for AG biosynthesis of Mycobacterium tuberculosis.
Gong Y, Wei C, Wang J, Mu N, Lu Q, Wu C, Yan N, Yang H, Zhao Y, Yang X, Gurcha SS, Veerapen N, Batt SM, Hao Z, Da L, Besra GS, Rao Z, Zhang L. Gong Y, et al. Proc Natl Acad Sci U S A. 2023 Jun 6;120(23):e2302858120. doi: 10.1073/pnas.2302858120. Epub 2023 May 30. Proc Natl Acad Sci U S A. 2023. PMID: 37252995 Free PMC article. - Characterization of Arabinosyl Transfer Reactions in the Biosynthesis of Mycobacterial Cell Envelope (Lipo)Polysaccharides.
Angala SK, Jackson M. Angala SK, et al. Methods Mol Biol. 2019;1954:175-186. doi: 10.1007/978-1-4939-9154-9_14. Methods Mol Biol. 2019. PMID: 30864132 Free PMC article. - The Mycobacterial Cell Wall--Peptidoglycan and Arabinogalactan.
Alderwick LJ, Harrison J, Lloyd GS, Birch HL. Alderwick LJ, et al. Cold Spring Harb Perspect Med. 2015 Mar 27;5(8):a021113. doi: 10.1101/cshperspect.a021113. Cold Spring Harb Perspect Med. 2015. PMID: 25818664 Free PMC article. Review. - Use of Synthetic Glycolipids to Probe the Number and Position of Arabinan Chains on Mycobacterial Arabinogalactan.
Angala SK, Joe M, McNeil MR, Liav A, Lowary TL, Jackson M. Angala SK, et al. ACS Chem Biol. 2021 Jan 15;16(1):20-26. doi: 10.1021/acschembio.0c00765. Epub 2020 Dec 31. ACS Chem Biol. 2021. PMID: 33382235 Free PMC article.
References
- Alderwick, L. J., M. Seidel, H. Sahm, G. S. Besra, and L. Eggeling. 2006. Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 28115653-15661. - PubMed
- Belanger, A. E., G. S. Besra, M. E. Ford, K. Mikusova, J. T. Belisle, P. J. Brennan, and J. M. Inamine. 1996. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc. Natl. Acad. Sci. USA 9311919-11924. - PMC - PubMed
- Belanger, A. E., and J. I. Inamine. 2000. Genetics of cell wall biosynthesis, p. 191-202. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, DC.
- Berg, S., D. Kaur, M. Jackson, and P. J. Brennan. 2007. The glycosyltransferases of Mycobacterium tuberculosis; roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology 1735R-56R. - PubMed
- Berg, S., J. Starbuck, J. B. Torrelles, V. D. Vissa, D. C. Crick, D. Chatterjee, and P. J. Brennan. 2005. Roles of conserved proline and glycosyltransferase motifs of EmbC in biosynthesis of lipoarabinomannan. J. Biol. Chem. 2805651-5663. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous