MicroRNA expression and identification of putative miRNA targets in ovarian cancer - PubMed (original) (raw)
MicroRNA expression and identification of putative miRNA targets in ovarian cancer
Neetu Dahiya et al. PLoS One. 2008.
Abstract
Background: MicroRNAs (miRNAs) represent a class of small non-coding RNAs that control gene expression by targeting mRNAs and triggering either translation repression or RNA degradation. Emerging evidence suggests the potential involvement of altered regulation of miRNA in the pathogenesis of cancers, and these genes are thought to function as both tumor suppressors and oncogenes.
Methodology/principal findings: Using microRNA microarrays, we identify several miRNAs aberrantly expressed in human ovarian cancer tissues and cell lines. miR-221 stands out as a highly elevated miRNA in ovarian cancer, while miR-21 and several members of the let-7 family are found downregulated. Public databases were used to reveal potential targets for the highly differentially expressed miRNAs. In order to experimentally identify transcripts whose stability may be affected by the differentially expressed miRNAs, we transfected precursor miRNAs into human cancer cell lines and used oligonucleotide microarrays to examine changes in the mRNA levels. Interestingly, there was little overlap between the predicted and the experimental targets or pathways, or between experimental targets/pathways obtained using different cell lines, highlighting the complexity of miRNA target selection.
Conclusion/significance: Our results identify several differentially expressed miRNAs in ovarian cancer and identify potential target transcripts that may be regulated by these miRNAs. These miRNAs and their targets may have important roles in the initiation and development of ovarian cancer.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
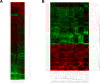
Figure 1. Cluster analysis of miRNA expression.
Tree generated by cluster analysis of ovarian cancer tissues and cell lines based on (A) all tested miRNAs in tissues and cell lines, and (B) differentially regulated miRNAs (Fold change >2.0 or <0.5 in greater than 60% of the samples) in tissues and cell lines compared to the normal control HOSE-B cells.
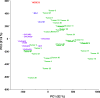
Figure 2. Principal component analysis of ovarian cancer samples (PCA) based on global miRNA expression.
Two-dimensional PCA shows that global miRNA expression patterns are different in ovarian cancer cell lines (indicated in blue), ovarian cancer tissues (indicated in green), and the non-tumorigenic HOSE-B cells (in red).
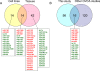
Figure 3. Comparisons of miRNA expression in ovarian tissues.
(A) The Venn diagram shows the number miRNAs differentially expressed in ovarian cell lines, in ovarian cancer tissues and in both. For each category, the miRNAs elevated (indicated in red) and downregulated (indicated in green) are indicated below the diagram. (B) The Venn diagram shows the number of differentially expressed miRNAs identified in the current study and the number of miRNAs indentified in 3 previous ovarian cancer studies. The miRNAs in common are indicated below the diagram and color-coded (red: elevated; green: decreased).
Figure 4. Forced overexpression of selected miRNAs in ovarian cancer cell lines.
Pre-miR-34c, Pre-miR-98, Pre-miR-424, Pre-let-7f were overexpressed in BG-1 and UCI-101. The products for each of the miRNAs is shown in duplicate for the two cell lines used. Significant overexpression of the miRNAs is confirmed. RT-PCR of 18S RNA is shown for each condition to demonstrate equal loading.
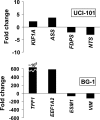
Figure 5. Validation of illumina arrays data for let-7f.
Transcripts identified by illumina arrays to be altered following let-7f overexpression are validated by RT-PCR. Fold changes for genes KIF1A, ASS, FDPS, NTS (in UCI-101 cells), and TFF1, EEF1A2, ESM1, VIM (in BG-1 cells) are shown and confirm the changes identified by illumina arrays.
Similar articles
- A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer.
Nagaraja AK, Creighton CJ, Yu Z, Zhu H, Gunaratne PH, Reid JG, Olokpa E, Itamochi H, Ueno NT, Hawkins SM, Anderson ML, Matzuk MM. Nagaraja AK, et al. Mol Endocrinol. 2010 Feb;24(2):447-63. doi: 10.1210/me.2009-0295. Epub 2010 Jan 15. Mol Endocrinol. 2010. PMID: 20081105 Free PMC article. - MicroRNA signatures in human ovarian cancer.
Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Ménard S, Croce CM. Iorio MV, et al. Cancer Res. 2007 Sep 15;67(18):8699-707. doi: 10.1158/0008-5472.CAN-07-1936. Cancer Res. 2007. PMID: 17875710 - miRNA profiling along tumour progression in ovarian carcinoma.
Vaksman O, Stavnes HT, Kaern J, Trope CG, Davidson B, Reich R. Vaksman O, et al. J Cell Mol Med. 2011 Jul;15(7):1593-602. doi: 10.1111/j.1582-4934.2010.01148.x. J Cell Mol Med. 2011. PMID: 20716115 Free PMC article. - MicroRNAs in ovarian carcinomas.
Dahiya N, Morin PJ. Dahiya N, et al. Endocr Relat Cancer. 2010 Jan 29;17(1):F77-89. doi: 10.1677/ERC-09-0203. Print 2010 Mar. Endocr Relat Cancer. 2010. PMID: 19903743 Free PMC article. Review. - Role of microRNAs in gynecological pathology.
Gilabert-Estelles J, Braza-Boils A, Ramon LA, Zorio E, Medina P, Espana F, Estelles A. Gilabert-Estelles J, et al. Curr Med Chem. 2012;19(15):2406-13. doi: 10.2174/092986712800269362. Curr Med Chem. 2012. PMID: 22455593 Review.
Cited by
- The Functional Role of the Long Non-Coding RNA LINCMD1 in Leiomyoma Pathogenesis.
Chuang TD, Ton N, Rysling S, Khorram O. Chuang TD, et al. Int J Mol Sci. 2024 Oct 27;25(21):11539. doi: 10.3390/ijms252111539. Int J Mol Sci. 2024. PMID: 39519092 Free PMC article. - The role of circular RNA targeting IGF2BPs in cancer-a potential target for cancer therapy.
Luo X, Shi J, Wang S, Jin X. Luo X, et al. J Mol Med (Berl). 2024 Nov;102(11):1297-1314. doi: 10.1007/s00109-024-02488-8. Epub 2024 Sep 17. J Mol Med (Berl). 2024. PMID: 39287635 Review. - LncRNA HOXA‑AS2 promotes the progression of epithelial ovarian cancer via the regulation of miR‑372.
Wang Y, Shao W. Wang Y, et al. Oncol Lett. 2024 Jun 25;28(2):394. doi: 10.3892/ol.2024.14527. eCollection 2024 Aug. Oncol Lett. 2024. PMID: 38966577 Free PMC article. - Novel DNA methylation-based epigenetic signatures in colorectal cancer from peripheral blood leukocytes.
Jung SY, Yu H, Tan X, Pellegrini M. Jung SY, et al. Am J Cancer Res. 2024 May 15;14(5):2253-2271. doi: 10.62347/MXWJ1398. eCollection 2024. Am J Cancer Res. 2024. PMID: 38859857 Free PMC article. - The microRNA Let-7 and its exosomal form: Epigenetic regulators of gynecological cancers.
Wang F, Zhou C, Zhu Y, Keshavarzi M. Wang F, et al. Cell Biol Toxicol. 2024 Jun 5;40(1):42. doi: 10.1007/s10565-024-09884-3. Cell Biol Toxicol. 2024. PMID: 38836981 Free PMC article. Review.
References
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. - PubMed
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. - PubMed
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Vasudevan S, Tong Y, Steitz JA. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science. 2007 - PubMed
- Tran N, McLean T, Zhang X, Zhao CJ, Thomson JM, et al. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–17. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical