Spidermonkey: rapid detection of co-evolving sites using Bayesian graphical models - PubMed (original) (raw)
Spidermonkey: rapid detection of co-evolving sites using Bayesian graphical models
Art F Y Poon et al. Bioinformatics. 2008.
Abstract
Spidermonkey is a new component of the Datamonkey suite of phylogenetic tools that provides methods for detecting coevolving sites from a multiple alignment of homologous nucleotide or amino acid sequences. It reconstructs the substitution history of the alignment by maximum likelihood-based phylogenetic methods, and then analyzes the joint distribution of substitution events using Bayesian graphical models to identify significant associations among sites.
Availability: Spidermonkey is publicly available both as a web application at http://www.data-monkey.org and as a stand-alone component of the phylogenetic software package HyPhy, which is freely distributed on the web (http://www.hyphy.org) as precompiled binaries and open source.
Figures
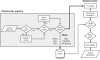
Fig. 1.
Flowchart diagram of the Spidermonkey pipeline. Abbreviations: SLAC=single likelihood ancestor counting; FEL=fixed effects likelihood; IFEL=internal FEL; REL=random effects likelihood (Kosakovsky Pond and Frost, 2005c); PARRIS=a partitioning approach for robust inference of selection (Scheffler et al., 2006); GA-Branch=genetic algorithm for detecting branch-specific selection (Kosakovsky Pond and Frost, 2005b).
Similar articles
- Detecting Amino Acid Coevolution with Bayesian Graphical Models.
Avino M, Poon AFY. Avino M, et al. Methods Mol Biol. 2019;1851:105-122. doi: 10.1007/978-1-4939-8736-8_6. Methods Mol Biol. 2019. PMID: 30298394 - Datamonkey: rapid detection of selective pressure on individual sites of codon alignments.
Pond SL, Frost SD. Pond SL, et al. Bioinformatics. 2005 May 15;21(10):2531-3. doi: 10.1093/bioinformatics/bti320. Epub 2005 Feb 15. Bioinformatics. 2005. PMID: 15713735 - Malin: maximum likelihood analysis of intron evolution in eukaryotes.
Csurös M. Csurös M. Bioinformatics. 2008 Jul 1;24(13):1538-9. doi: 10.1093/bioinformatics/btn226. Epub 2008 May 12. Bioinformatics. 2008. PMID: 18474506 Free PMC article. - Multiple sequence alignments.
Wallace IM, Blackshields G, Higgins DG. Wallace IM, et al. Curr Opin Struct Biol. 2005 Jun;15(3):261-6. doi: 10.1016/j.sbi.2005.04.002. Curr Opin Struct Biol. 2005. PMID: 15963889 Review. - Finding homologs to nucleic acid or protein sequences using the framesearch program.
Healy M. Healy M. Curr Protoc Bioinformatics. 2002 Aug;Chapter 3:Unit 3.2. doi: 10.1002/0471250953.bi0302s00. Curr Protoc Bioinformatics. 2002. PMID: 18792937 Review.
Cited by
- Evolutionary analysis of ZAP and its cofactors identifies intrinsically disordered regions as central elements in host-pathogen interactions.
Cagliani R, Forni D, Mozzi A, Fuchs R, Hagai T, Sironi M. Cagliani R, et al. Comput Struct Biotechnol J. 2024 Aug 2;23:3143-3154. doi: 10.1016/j.csbj.2024.07.022. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39234301 Free PMC article. - The evolution of mammalian Rem2: unraveling the impact of purifying selection and coevolution on protein function, and implications for human disorders.
Lucaci AG, Brew WE, Lamanna J, Selberg A, Carnevale V, Moore AR, Kosakovsky Pond SL. Lucaci AG, et al. Front Bioinform. 2024 Jun 24;4:1381540. doi: 10.3389/fbinf.2024.1381540. eCollection 2024. Front Bioinform. 2024. PMID: 38978817 Free PMC article. - AOC: Analysis of Orthologous Collections - an application for the characterization of natural selection in protein-coding sequences.
Lucaci AG, Pond SLK. Lucaci AG, et al. ArXiv [Preprint]. 2024 Jun 13:arXiv:2406.09522v1. ArXiv. 2024. PMID: 38947939 Free PMC article. Preprint. - Genetic Variation and Evolutionary Analysis of Eggplant Mottled Dwarf Virus Isolates from Spain.
Alfaro-Fernández A, Taengua R, Font-San-Ambrosio I, Sanahuja-Edo E, Peiró R, Galipienso L, Rubio L. Alfaro-Fernández A, et al. Plants (Basel). 2024 Jan 16;13(2):250. doi: 10.3390/plants13020250. Plants (Basel). 2024. PMID: 38256804 Free PMC article. - HIV-1 Group M Capsid Amino Acid Variability: Implications for Sequence Quality Control of Genotypic Resistance Testing.
Tao K, Rhee SY, Tzou PL, Osman ZA, Pond SLK, Holmes SP, Shafer RW. Tao K, et al. Viruses. 2023 Apr 18;15(4):992. doi: 10.3390/v15040992. Viruses. 2023. PMID: 37112972 Free PMC article.
References
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 1985;125:1–15.
- Friedman N, Koller D. Being Bayesian about network structure. A Bayesian approach to structure discovery in Bayesian networks. Mach. Learn. 2003;50:95–125.
- Gansner E, North S. Software: Practice and Experience. Chichester, New York: John Wiley & Sons, Ltd.; 2000. An open graph visualization system and its applications to software engineering.
- Kosakovsky Pond SL, Frost S.DW. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005a;21:2531–2533. - PubMed
- Kosakovsky Pond SL, Frost S.DW. A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol. Biol. Evol. 2005b;22:478–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI057167/AI/NIAID NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AI047745/AI/NIAID NIH HHS/United States
- AI36214/AI/NIAID NIH HHS/United States
- R21 AI047745/AI/NIAID NIH HHS/United States
- U01 AI043638/AI/NIAID NIH HHS/United States
- R56 AI047745/AI/NIAID NIH HHS/United States
- P30 AI036214/AI/NIAID NIH HHS/United States
- AI57167/AI/NIAID NIH HHS/United States
- AI43638/AI/NIAID NIH HHS/United States
- AI47745/AI/NIAID NIH HHS/United States