Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide - PubMed (original) (raw)
Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide
Berend Jan Bosch et al. J Virol. 2008 Sep.
Abstract
Unlike other class I viral fusion proteins, spike proteins on severe acute respiratory syndrome coronavirus virions are uncleaved. As we and others have demonstrated, infection by this virus depends on cathepsin proteases present in endosomal compartments of the target cell, suggesting that the spike protein acquires its fusion competence by cleavage during cell entry rather than during virion biogenesis. Here we demonstrate that cathepsin L indeed activates the membrane fusion function of the spike protein. Moreover, cleavage was mapped to the same region where, in coronaviruses carrying furin-activated spikes, the receptor binding subunit of the protein is separated from the membrane-anchored fusion subunit.
Figures
FIG. 1.
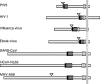
Schematic diagrams of the fusion proteins of parainfluenza virus 5 (PIV5; formerly known as simian virus 5), human immunodeficiency virus type 1 (HIV-1), influenza virus, Ebola virus, SARS-CoV (strain BJ01), MHV-A59, and HCoV-NL63. Filled bars represent the fusion peptide. Dark and light shaded bars, HR1 and HR2 regions, respectively. Open arrowheads indicate the positions of the furin cleavage sites. The fusion proteins are C-terminally anchored in the viral membrane (long vertical shaded bar).
FIG. 2.
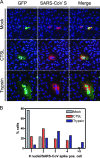
(A) Confocal fluorescence microscopy of Vero cells transfected with plasmids expressing the SARS-CoV spike or GFP gene and either mock treated or treated with cathepsin L protease (CTSL; 2 μg/ml) or trypsin (2 μg/ml). Nuclei (blue) were labeled with ToPro-3 (Molecular Probes). (B) Semiquantitation of syncytium formation. The number of nuclei in SARS-CoV spike protein-expressing cells/syncytia was counted in mock-treated, CTSL (2 μg/ml)-treated, and trypsin (2 μg/ml)-treated Vero cell cultures. pos., positive.
FIG. 3.
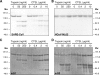
Cathepsin L (CTSL) cleavage of the SARS-CoV spike protein. Purified trimeric spike ectodomains of SARS-CoV and HCoV-NL63 were incubated with different concentrations of CTSL or TPCK-treated trypsin for 30 min at room temperature. (A and B) Samples were analyzed by SDS-PAGE and stained with GelCode Blue reagent. (C and D) SARS-CoV samples were also subjected to Western blot analysis using antibodies recognizing the N-terminal part (α-S1) or the C-terminal part (α-S2) of the SARS-CoV spike ectodomain, respectively. The positions of molecular weight (Mw) marker proteins (in thousands) are indicated alongside the gels and blots.
FIG. 4.
Sequence alignment of the SARS-CoV (strain BJ01; GenBank accession no. AY278488) and MHV-A59 (primary accession no. P11224) spike proteins at the S1-S2 junction. Identical residues are asterisked. The filled, shaded, and open arrowheads indicate the cathepsin L, trypsin, and furin cleavage sites, respectively, with their cleavage positions given in parentheses.
Similar articles
- Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry.
Qiu Z, Hingley ST, Simmons G, Yu C, Das Sarma J, Bates P, Weiss SR. Qiu Z, et al. J Virol. 2006 Jun;80(12):5768-76. doi: 10.1128/JVI.00442-06. J Virol. 2006. PMID: 16731916 Free PMC article. - Proteolysis of SARS-associated coronavirus spike glycoprotein.
Simmons G, Rennekamp AJ, Bates P. Simmons G, et al. Adv Exp Med Biol. 2006;581:235-40. doi: 10.1007/978-0-387-33012-9_39. Adv Exp Med Biol. 2006. PMID: 17037535 Free PMC article. No abstract available. - SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells.
Huang IC, Bosch BJ, Li F, Li W, Lee KH, Ghiran S, Vasilieva N, Dermody TS, Harrison SC, Dormitzer PR, Farzan M, Rottier PJ, Choe H. Huang IC, et al. J Biol Chem. 2006 Feb 10;281(6):3198-203. doi: 10.1074/jbc.M508381200. Epub 2005 Dec 8. J Biol Chem. 2006. PMID: 16339146 Free PMC article. - A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L.
Pager CT, Craft WW Jr, Patch J, Dutch RE. Pager CT, et al. Virology. 2006 Mar 15;346(2):251-7. doi: 10.1016/j.virol.2006.01.007. Epub 2006 Feb 7. Virology. 2006. PMID: 16460775 Free PMC article.
Cited by
- Phosphatidylinositol 4-kinase IIIβ is required for severe acute respiratory syndrome coronavirus spike-mediated cell entry.
Yang N, Ma P, Lang J, Zhang Y, Deng J, Ju X, Zhang G, Jiang C. Yang N, et al. J Biol Chem. 2012 Mar 9;287(11):8457-67. doi: 10.1074/jbc.M111.312561. Epub 2012 Jan 17. J Biol Chem. 2012. PMID: 22253445 Free PMC article. - SARS-CoV-2 receptor ACE2 protein expression in serum is significantly associated with age.
Pavel AB, Wu J, Renert-Yuval Y, Del Duca E, Glickman JW, Miller RL, Paller AS, Krueger JG, Guttman-Yassky E. Pavel AB, et al. Allergy. 2021 Mar;76(3):875-878. doi: 10.1111/all.14522. Epub 2020 Aug 24. Allergy. 2021. PMID: 32726474 Free PMC article. No abstract available. - Leaving no stone unturned: Allosteric targeting of SARS-CoV-2 spike protein at putative druggable sites disrupts human angiotensin-converting enzyme interactions at the receptor binding domain.
Olotu FA, Omolabi KF, Soliman MES. Olotu FA, et al. Inform Med Unlocked. 2020;21:100451. doi: 10.1016/j.imu.2020.100451. Epub 2020 Oct 16. Inform Med Unlocked. 2020. PMID: 33083517 Free PMC article. - A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry.
Wong AC, Sandesara RG, Mulherkar N, Whelan SP, Chandran K. Wong AC, et al. J Virol. 2010 Jan;84(1):163-75. doi: 10.1128/JVI.01832-09. J Virol. 2010. PMID: 19846533 Free PMC article. - Middle East Respiratory Syndrome Coronavirus Spike Protein Is Not Activated Directly by Cellular Furin during Viral Entry into Target Cells.
Matsuyama S, Shirato K, Kawase M, Terada Y, Kawachi K, Fukushi S, Kamitani W. Matsuyama S, et al. J Virol. 2018 Sep 12;92(19):e00683-18. doi: 10.1128/JVI.00683-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30021905 Free PMC article.
References
- Bosch, B. J., B. E. Martina, R. Van Der Zee, J. Lepault, B. J. Haijema, C. Versluis, A. J. Heck, R. De Groot, A. D. Osterhaus, and P. J. Rottier. 2004. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA 1018455-8460. - PMC - PubMed
- Calder, L. J., L. Gonzalez-Reyes, B. Garcia-Barreno, S. A. Wharton, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2000. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology 271122-131. - PubMed
- Chambers, P., C. R. Pringle, and A. J. Easton. 1990. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 713075-3080. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials