Assembly reflects evolution of protein complexes - PubMed (original) (raw)
. 2008 Jun 26;453(7199):1262-5.
doi: 10.1038/nature06942. Epub 2008 Jun 18.
Affiliations
- PMID: 18563089
- PMCID: PMC2658002
- DOI: 10.1038/nature06942
Assembly reflects evolution of protein complexes
Emmanuel D Levy et al. Nature. 2008.
Abstract
A homomer is formed by self-interacting copies of a protein unit. This is functionally important, as in allostery, and structurally crucial because mis-assembly of homomers is implicated in disease. Homomers are widespread, with 50-70% of proteins with a known quaternary state assembling into such structures. Despite their prevalence, their role in the evolution of cellular machinery and the potential for their use in the design of new molecular machines, little is known about the mechanisms that drive formation of homomers at the level of evolution and assembly in the cell. Here we present an analysis of over 5,000 unique atomic structures and show that the quaternary structure of homomers is conserved in over 70% of protein pairs sharing as little as 30% sequence identity. Where quaternary structure is not conserved among the members of a protein family, a detailed investigation revealed well-defined evolutionary pathways by which proteins transit between different quaternary structure types. Furthermore, we show by perturbing subunit interfaces within complexes and by mass spectrometry analysis, that the (dis)assembly pathway mimics the evolutionary pathway. These data represent a molecular analogy to Haeckel's evolutionary paradigm of embryonic development, where an intermediate in the assembly of a complex represents a form that appeared in its own evolutionary history. Our model of self-assembly allows reliable prediction of evolution and assembly of a complex solely from its crystal structure.
Figures
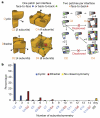
Figure 1. Abundance and properties of cyclic and dihedral symmetries
a, n subunits in a cyclic complex are related by a single _n_-fold symmetry axis (dotted lines); ellipses and squares represent two- and four-fold axes, respectively. For a monomer to evolve towards a cyclic tetramer (C4), two complementary surfaces have to evolve simultaneously (red and orange patches). For a dihedral tetramer (D2), two different and self-complementary surfaces (green and blue patches) can evolve serially with an intermediate dimer (C2). b, The abundance of homomers with cyclic, dihedral, or no symmetry (3.5%). 62.7% are cyclic dimers (C2), 8% are cyclic trimers (C3), and 3.2% have higher order cyclic symmetry (from C4 to C14). Dihedral complexes dominate (22.6%) among complexes with ≥4 subunits.
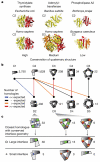
Figure 2. Routes for homomer evolution
a, Illustrative examples of different levels of quaternary structure conservation (termed high, medium and low). Thymidylate synthases (PDB accessions 1ajm and 1hw3) are both dimers. Adenylyltransferases (PDB accessions 1kam and 1kr2) differ in their number of subunits, with similar dimers (yellow) common to both quaternary structures. The dimer and trimer of venom-toxin phospholipase A2s are not related geometrically and the quaternary structures are therefore less conserved. b, Schematic illustration of large-scale analysis of quaternary structures; the number of unique complexes is indicated. Coloured lines indicate the significance in over-representation of shared homologous complexes (Methods). c, In 49 out of 52 cases, the largest interface is present in the dimeric or trimeric homologue, illustrated by size of interface patches.
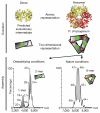
Figure 3. Prediction of evolutionary routes and link with (dis)assembly in solution
ATP sulphurylase is a hexamer in Penicillium chrysogenum, with a predicted dimeric evolutionary intermediate based on interface sizes (that is, the interface in the dimer (green patch) is larger than the trimeric interface (red and orange patch; top panel)). We perturb the hexameric ATP sulphurylase (PDB accession 1m8p, Supplementary Table 2) to disassemble it into subcomplexes probed using electrospray mass spectrometry (bottom panel) and determine whether a dimeric (with the larger interface) or a trimeric (small interface) subcomplex is detected. Both dimers and monomers were detected, with no evidence of trimers.
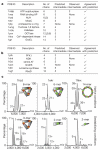
Figure 4. (Dis)assembly pathways in 16 complexes
a, Homomers for which (dis)assembly was probed using electrospray mass spectrometry (Methods). ‘None’ denotes no intermediate detected. Complexes agree with our prediction where the subcomplex containing the larger interface is the most stable in eight out of ten cases. b, List of homomers for which: (1) information on (dis)assembly has been reported (Supplementary Table 3); (2) a crystal structure is known; and (3) the intermediate species observed during (dis)assembly could be mapped to a subcomplex in the structure. For these complexes, we found an agreement with our prediction in 5/6 cases. c, Mass spectra showing intact complexes (top panel) as well as subcomplexes obtained after destabilization in solution (bottom panel). AUH, an RNA binding protein; GroEL and GroES are chaperonins; KHMase, ketopantoate hydroxymethyltransferase; MoaC, molybdenum cofactor biosynthesis protein; RNA binding AP, RNA binding antitermination protein; serine AT, serine acyltransferase; OCTase, ornithine carbamoyltransferase.
Similar articles
- Structural, evolutionary, and assembly principles of protein oligomerization.
Levy ED, Teichmann S. Levy ED, et al. Prog Mol Biol Transl Sci. 2013;117:25-51. doi: 10.1016/B978-0-12-386931-9.00002-7. Prog Mol Biol Transl Sci. 2013. PMID: 23663964 Review. - Structure, dynamics, assembly, and evolution of protein complexes.
Marsh JA, Teichmann SA. Marsh JA, et al. Annu Rev Biochem. 2015;84:551-75. doi: 10.1146/annurev-biochem-060614-034142. Epub 2014 Dec 8. Annu Rev Biochem. 2015. PMID: 25494300 Review. - Ligand Binding Site Structure Shapes Folding, Assembly and Degradation of Homomeric Protein Complexes.
Abrusán G, Marsh JA. Abrusán G, et al. J Mol Biol. 2019 Sep 6;431(19):3871-3888. doi: 10.1016/j.jmb.2019.07.014. Epub 2019 Jul 12. J Mol Biol. 2019. PMID: 31306664 Free PMC article. - Protein complexes are under evolutionary selection to assemble via ordered pathways.
Marsh JA, Hernández H, Hall Z, Ahnert SE, Perica T, Robinson CV, Teichmann SA. Marsh JA, et al. Cell. 2013 Apr 11;153(2):461-70. doi: 10.1016/j.cell.2013.02.044. Cell. 2013. PMID: 23582331 Free PMC article. - The emergence of protein complexes: quaternary structure, dynamics and allostery. Colworth Medal Lecture.
Perica T, Marsh JA, Sousa FL, Natan E, Colwell LJ, Ahnert SE, Teichmann SA. Perica T, et al. Biochem Soc Trans. 2012 Jun 1;40(3):475-91. doi: 10.1042/BST20120056. Biochem Soc Trans. 2012. PMID: 22616857
Cited by
- Supercharged fluorescent proteins detect lanthanides via direct antennae signaling.
Huang KY, Cardenas L, Ellington AD, Walker DJF. Huang KY, et al. Nat Commun. 2024 Oct 24;15(1):9200. doi: 10.1038/s41467-024-53106-7. Nat Commun. 2024. PMID: 39448572 Free PMC article. - Cellular location shapes quaternary structure of enzymes.
Abrusán G, Zelezniak A. Abrusán G, et al. Nat Commun. 2024 Oct 1;15(1):8505. doi: 10.1038/s41467-024-52662-2. Nat Commun. 2024. PMID: 39353940 Free PMC article. - Evolution and maintenance of mtDNA gene content across eukaryotes.
Veeraragavan S, Johansen M, Johnston IG. Veeraragavan S, et al. Biochem J. 2024 Aug 7;481(15):1015-1042. doi: 10.1042/BCJ20230415. Biochem J. 2024. PMID: 39101615 Free PMC article. Review. - Symmetry facilitated the evolution of heterospecificity and high-order stoichiometry in vertebrate hemoglobin.
Cortez-Romero CR, Lyu J, Pillai AS, Laganowsky A, Thornton JW. Cortez-Romero CR, et al. bioRxiv [Preprint]. 2024 Dec 13:2024.07.24.604985. doi: 10.1101/2024.07.24.604985. bioRxiv. 2024. PMID: 39091803 Free PMC article. Preprint. - Frequent transitions in self-assembly across the evolution of a central metabolic enzyme.
Sendker FL, Schlotthauer T, Mais CN, Lo YK, Girbig M, Bohn S, Heimerl T, Schindler D, Weinstein A, Metzger BP, Thornton JW, Pillai A, Bange G, Schuller JM, Hochberg GKA. Sendker FL, et al. bioRxiv [Preprint]. 2024 Jul 7:2024.07.05.602260. doi: 10.1101/2024.07.05.602260. bioRxiv. 2024. PMID: 39005358 Free PMC article. Updated. Preprint.
References
- Cabezon E, et al. Homologous and heterologous inhibitory effects of ATPase inhibitor proteins on F-ATPases. J. Biol. Chem. 2002;277:41334–41341. - PubMed
- Hardy LW, et al. Atomic structure of thymidylate synthase: target for rational drug design. Science. 1987;235:448–455. - PubMed
- Iber D, Clarkson J, Yudkin MD, Campbell ID. The mechanism of cell differentiation in Bacillus subtilis. Nature. 2006;441:371–374. - PubMed
- Marianayagam NJ, Sunde M, Matthews JM. The power of two: protein dimerization in biology. Trends Biochem. Sci. 2004;29:618–625. - PubMed
- Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources