MITOMASTER: a bioinformatics tool for the analysis of mitochondrial DNA sequences - PubMed (original) (raw)
MITOMASTER: a bioinformatics tool for the analysis of mitochondrial DNA sequences
Marty C Brandon et al. Hum Mutat. 2009 Jan.
Abstract
We have developed a computer system, MITOMASTER, to make analysis of human mitochondrial DNA (mtDNA) sequences efficient, accurate, and easily available. From imported sequences, the system identifies nucleotide variants, determines the haplogroup, rules out possible pseudogene contamination, identifies novel DNA sequence variants, and evaluates the potential biological significance of each variant. This system should be beneficial for mtDNA analyses of biomedical physicians and investigators, population biologists and forensic scientists. MITOMASTER can be accessed at http://mammag.web.uci.edu/twiki/bin/view/Mitomaster.
Copyright 2008 Wiley-Liss, Inc.
Figures
Figure 1
The algorithm used by biologist, and implemented within MITOMASTER, to identify possible clinical variants within a patient's mtDNA sequence. Variants are extracted by alignment with a reference sequence; the group of extracted variants is used to determine the haplogroup of the sequence and each individual variant is checked within the database and analyzed to determine its biological effect; variation within coding loci is further analyzed to determine its coding effect and inter-species rate of conservation.
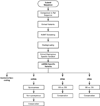
Figure 2
MITOMASTER's functional components. A website interface allows users to submit either files of whole mtDNA sequences or individual variants for analysis. Programs within the analysis component compare the submission with the mCRS, extract the variants found within submitted sequences, perform NUMT screening on the variants extracted, assign the sequences to a haplogroup, and determine the predicted functional effect of each variant.
Figure 3
Simplified schema diagram for MITOMASTER's database. DNA sequences are decomposed into their constituent nucleic acids and entries made in the dna table. During simulated transcription and translation processes, all variation (relative to the mCRS) is entered into the rna and aa tables, respectively. Some variation involves multiple base alterations and these cases are grouped in the “variant” table. The aggregated variants that define the known haplogroups are represented in the haplogroups data set, the nodes are the branch points of the mtDNA phylogenetic tree.
Similar articles
- Mitochondrial cardiomyopathies: how to identify candidate pathogenic mutations by mitochondrial DNA sequencing, MITOMASTER and phylogeny.
Zaragoza MV, Brandon MC, Diegoli M, Arbustini E, Wallace DC. Zaragoza MV, et al. Eur J Hum Genet. 2011 Feb;19(2):200-7. doi: 10.1038/ejhg.2010.169. Epub 2010 Oct 27. Eur J Hum Genet. 2011. PMID: 20978534 Free PMC article. - mtDNA Variation and Analysis Using Mitomap and Mitomaster.
Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, Procaccio V, Wallace DC. Lott MT, et al. Curr Protoc Bioinformatics. 2013 Dec;44(123):1.23.1-26. doi: 10.1002/0471250953.bi0123s44. Curr Protoc Bioinformatics. 2013. PMID: 25489354 Free PMC article. - HAPLOFIND: a new method for high-throughput mtDNA haplogroup assignment.
Vianello D, Sevini F, Castellani G, Lomartire L, Capri M, Franceschi C. Vianello D, et al. Hum Mutat. 2013 Sep;34(9):1189-94. doi: 10.1002/humu.22356. Epub 2013 Jun 12. Hum Mutat. 2013. PMID: 23696374 - An enhanced MITOMAP with a global mtDNA mutational phylogeny.
Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. Ruiz-Pesini E, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D823-8. doi: 10.1093/nar/gkl927. Epub 2006 Dec 18. Nucleic Acids Res. 2007. PMID: 17178747 Free PMC article. - SWORDS: a statistical tool for analysing large DNA sequences.
Chaudhuri P, Das S. Chaudhuri P, et al. J Biosci. 2002 Feb;27(1 Suppl 1):1-6. doi: 10.1007/BF02703678. J Biosci. 2002. PMID: 11927772 Review.
Cited by
- African-Colombian woman with preeclampsia and high-risk APOL1 genotype: A case report.
Duran CE, Gutierrez-Medina JD, Triviño Arias J, Sandoval-Calle LM, Barbosa M, Useche E, Diaz-Ordoñez L, Pachajoa H. Duran CE, et al. Medicine (Baltimore). 2024 Nov 1;103(44):e40284. doi: 10.1097/MD.0000000000040284. Medicine (Baltimore). 2024. PMID: 39496047 Free PMC article. - Whole-Exome Analysis for Polish Caucasian Patients with Retinal Dystrophies and the Creation of a Reference Genomic Database for the Polish Population.
Matczyńska E, Szymańczak R, Stradomska K, Łyszkiewicz P, Jędrzejowska M, Kamińska K, Beć-Gajowniczek M, Suchecka E, Zagulski M, Wiącek M, Wylęgała E, Machalińska A, Mossakowska M, Puzianowska-Kuźnicka M, Teper S, Boguszewska-Chachulska A. Matczyńska E, et al. Genes (Basel). 2024 Aug 1;15(8):1011. doi: 10.3390/genes15081011. Genes (Basel). 2024. PMID: 39202371 Free PMC article. - Mitochondrial DNA: Inherent Complexities Relevant to Genetic Analyses.
Ferreira T, Rodriguez S. Ferreira T, et al. Genes (Basel). 2024 May 12;15(5):617. doi: 10.3390/genes15050617. Genes (Basel). 2024. PMID: 38790246 Free PMC article. Review. - Mitochondrial DNA Variants at Low-Level Heteroplasmy and Decreased Copy Numbers in Chronic Kidney Disease (CKD) Tissues with Kidney Cancer.
Kanazashi Y, Maejima K, Johnson TA, Sasagawa S, Jikuya R, Hasumi H, Matsumoto N, Maekawa S, Obara W, Nakagawa H. Kanazashi Y, et al. Int J Mol Sci. 2023 Dec 7;24(24):17212. doi: 10.3390/ijms242417212. Int J Mol Sci. 2023. PMID: 38139039 Free PMC article. - Extraction Protocol for Parallel Analysis of Proteins and DNA from Ancient Teeth and Dental Calculus.
Chocholova E, Roudnicky P, Potesil D, Fialova D, Krystofova K, Drozdova E, Zdrahal Z. Chocholova E, et al. J Proteome Res. 2023 Oct 6;22(10):3311-3319. doi: 10.1021/acs.jproteome.3c00370. Epub 2023 Sep 12. J Proteome Res. 2023. PMID: 37699853 Free PMC article.
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. - PubMed
- Bannwarth S, Procaccio V, Paquis-Flucklinger V. Surveyor Nuclease: a new strategy for a rapid identification of heteroplasmic mitochondrial DNA mutations in patients with respiratory chain defects. Human Mutation. 2005;25(6):575–582. - PubMed
- Baudouin SV, Saunders D, Tiangyou W, Elson JL, Poynter J, Pyle A, Keers S, Turnbull DM, Howell N, Chinnery PF. Mitochondrial DNA and survival after sepsis: a prospective study. Lancet. 2005;366(9503):2118–2121. - PubMed
- Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25(34):4647–4662. - PubMed
- Brandon MC, Baldi P, Navathe SB, Lott MT, Wallace DC. Mitomaster - tool for diagnosing genetic diseases. Proceedings of the IEEE Computer Society Bioinformatics Conference. 2003
Publication types
MeSH terms
Substances
Grants and funding
- NS21328/NS/NINDS NIH HHS/United States
- DK73691/DK/NIDDK NIH HHS/United States
- AG24373/AG/NIA NIH HHS/United States
- R01 AG024373/AG/NIA NIH HHS/United States
- R01 DK073691/DK/NIDDK NIH HHS/United States
- R01 NS021328/NS/NINDS NIH HHS/United States
- R01 AG013154/AG/NIA NIH HHS/United States
- AG13154/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources