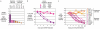IRF4 addiction in multiple myeloma - PubMed (original) (raw)
. 2008 Jul 10;454(7201):226-31.
doi: 10.1038/nature07064. Epub 2008 Jun 22.
N C Tolga Emre, Laurence Lamy, Vu N Ngo, George Wright, Wenming Xiao, John Powell, Sandeep Dave, Xin Yu, Hong Zhao, Yuxin Zeng, Bangzheng Chen, Joshua Epstein, Louis M Staudt
Affiliations
- PMID: 18568025
- PMCID: PMC2542904
- DOI: 10.1038/nature07064
IRF4 addiction in multiple myeloma
Arthur L Shaffer et al. Nature. 2008.
Abstract
The transcription factor IRF4 (interferon regulatory factor 4) is required during an immune response for lymphocyte activation and the generation of immunoglobulin-secreting plasma cells. Multiple myeloma, a malignancy of plasma cells, has a complex molecular aetiology with several subgroups defined by gene expression profiling and recurrent chromosomal translocations. Moreover, the malignant clone can sustain multiple oncogenic lesions, accumulating genetic damage as the disease progresses. Current therapies for myeloma can extend survival but are not curative. Hence, new therapeutic strategies are needed that target molecular pathways shared by all subtypes of myeloma. Here we show, using a loss-of-function, RNA-interference-based genetic screen, that IRF4 inhibition is toxic to myeloma cell lines, regardless of transforming oncogenic mechanism. Gene expression profiling and genome-wide chromatin immunoprecipitation analysis uncovered an extensive network of IRF4 target genes and identified MYC as a direct target of IRF4 in activated B cells and myeloma. Unexpectedly, IRF4 was itself a direct target of MYC transactivation, generating an autoregulatory circuit in myeloma cells. Although IRF4 is not genetically altered in most myelomas, they are nonetheless addicted to an aberrant IRF4 regulatory network that fuses the gene expression programmes of normal plasma cells and activated B cells.
Figures
Figure 1. IRF4 is required for myeloma cell survival
a, Cell lines were screened using a retrovirally-delivered, doxycycline-inducible, shRNA library to identify genes required for cell growth or survival, as described. Depletion of cells bearing an IRF4-targeted shRNA in shRNA-induced versus uninduced cells is plotted; error bars represent the s.d. of triplicate measurements. b, Expression of the IRF4 coding region rescues myeloma cells from lethality of an shRNA targeting the IRF4 3'UTR (see text for details). c, An IRF4 shRNA is toxic to myeloma but not lymphoma cell lines. A vector for constitutive expression of IRF4 shRNA was transduced into cell lines, and viability of shRNA+ cells was monitored. In (b) and (c), cells expressing shRNA were monitored by flow cytometry for a co-expressed GFP marker and data were normalized to the % of GFP+ cells at day 2 post infection.
Figure 2. IRF4 target genes in multiple myeloma
a, Venn diagram depicting IRF4 target genes and the overlap between the myeloma, plasma cell, and activated B cell gene expression programs. Of the 308 IRF4 target genes (Supplemental Fig. 3), 262 were well-measured on Affymetrix gene expression arrays. 101 were more highly expressed in primary myeloma samples than primary mature B cells (>1.4-fold, red circle), 67 were more highly expressed in primary plasma cells than mature B cells (>1.4-fold, green circle), and 81 are induced between 1 hr and 24 hr following activation of primary human B cells by anti-IgM crosslinking (>2-fold, yellow circle). red: direct IRF4 targets by ChIP, *: direct MYC targets. b, Representative conventional ChIP assays for genes identified as IRF4 targets by both gene expression profiling and ChIP-CHIP. Individual ChIP assays were performed on chromatin from the KMS12 myeloma line and the OCI-Ly19 lymphoma line using either anti-IRF4 or control antibodies. The ChIP signal is given in arbitrary relative units calculated from quantitative PCR data, based on the relative abundance of the indicated gene in the immunoprecipitated DNA versus input DNA. Error bars are s.d. from triplicate measurements.
Figure 3. MYC is a direct IRF4 target gene in myeloma and activated B cells
a, Knockdown of IRF4 decreases MYC mRNA expression. The SKMM1 myeloma line was transduced with IRF4 or MYC shRNAs, and gene expression was measured by quantitative RT-PCR after 4 days of shRNA induction, normalized to the signal from uninduced cells. b, Binding of IRF4 to the MYC promoter. ChIP was performed as in Figure 2, comparing the myeloma line KMS12 to the lymphoma line OCI-LY19, for regions of the MYC promoter (as indicated relative to the transcriptional start site) or a control locus, CYP2E1. c, IRF4 knockdown decreases IRF4 binding to the MYC promoter. ChIP was performed using KMS12 cells with or without shRNA induction for 4 days. d, Activation of human blood B cells leads to IRF4 binding to the MYC promoter. ChIP assays were performed using purified peripheral human blood B cells, either unstimulated or activated with P/I for the indicated times. e, Genetic deficiency of IRF4 impairs MYC induction during lymphocyte activation. Quantitative RT-PCR was performed on RNA from resting splenic B cells of IRF4-deficient or wild type mice, either unstimulated or activated with P/I for the indicated times. Values were normalized to B2M expression. f, IRF4 transactivates the MYC promoter. The OCI-Ly7 lymphoma line was transiently transfected with a GFP expression vector driven by the human MYC promoter, either alone, with an IRF4 expression vector, or with an empty vector control. Flow cytometry for GFP fluorescence is shown, with error bars indicating s.d. of triplicate measurements.
Figure 4. IRF4 is a direct MYC target gene in myeloma and activated B cells
a, Lethality of a MYC shRNA for cell lines expressing MYC. Cell lines were transduced with a MYC shRNA vector and the fraction of shRNA+ (GFP+) cells was monitored over time. All lines express MYC, except U266, which expresses MYCL1. b, Expression of the MYC coding region rescues H929 myeloma cells from lethality of an shRNA targeting the MYC 3'UTR. c, MYC knockdown downregulates MYC direct target genes and IRF4 target genes. KMS12 myeloma cells were induced for MYC shRNA expression for 4 days and profiled for gene expression changes. Each experiment utilized a different MYC shRNA. Exemplar array elements are shown (reduced by >1.3-fold in both experiments), for known MYC direct targets and IRF4 targets (this work). d, MYC knockdown decreases IRF4 mRNA expression. Shown are quantitative RT-PCR measurements of MYC and IRF4 mRNA levels in KMS12 myeloma cells, with or without induction of MYC shRNA. Error bars indicate s.d. of triplicate measurements. e, MYC binds to the IRF4 locus. ChIP of MYC binding to the IRF4 first intron in myeloma cells expressing MYC (KMS12, H929), but not in the myeloma line U266 that lacks MYC expression. f, MYC binding to the IRF4 locus is induced in activated human B cells. ChIP of MYC binding to the IRF4 first intron in human blood B cells, either unstimulated or activated with P/I for 6 hr. g, MYC and IRF4 are more highly expressed in primary myeloma patient samples than in normal human bone marrow plasma cells. Previously published gene expression profiling data was analyzed for mRNA expression of MYC, IRF4, and a control gene, UBC.
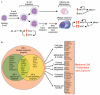
Figure 5. Model of IRF4 control over B cell development and multiple myeloma oncogenesis
a, IRF4 and MYC form a positive autoregulatory loop during normal B cell activation and in multiple myeloma. Genetic abnormalities of MYC upregulate its expression in myeloma, thereby augmenting IRF4 expression. In normal plasma cells, Blimp-1 represses MYC, but this control circuit may be abrogated in myeloma. b, IRF4 as a master regulator of the myeloma phenotype. IRF4 controls a myeloma-specific gene expression program that fuses the IRF4 regulatory programs from activated B cells and plasma cells. IRF4 direct targets regulate many essential cellular processes, causing myeloma cells to be addicted to IRF4.
Comment in
- Cancer: An unexpected addiction.
Shaughnessy JD. Shaughnessy JD. Nature. 2008 Jul 10;454(7201):172-3. doi: 10.1038/454172a. Nature. 2008. PMID: 18615074 No abstract available.
Similar articles
- Cancer: An unexpected addiction.
Shaughnessy JD. Shaughnessy JD. Nature. 2008 Jul 10;454(7201):172-3. doi: 10.1038/454172a. Nature. 2008. PMID: 18615074 No abstract available. - Dissecting the impact of bromodomain inhibitors on the Interferon Regulatory Factor 4-MYC oncogenic axis in multiple myeloma.
Agnarelli A, Mitchell S, Caalim G, Wood CD, Milton-Harris L, Chevassut T, West MJ, Mancini EJ. Agnarelli A, et al. Hematol Oncol. 2022 Aug;40(3):417-429. doi: 10.1002/hon.3016. Epub 2022 May 18. Hematol Oncol. 2022. PMID: 35544413 Free PMC article. - IRF4 in multiple myeloma-Biology, disease and therapeutic target.
Agnarelli A, Chevassut T, Mancini EJ. Agnarelli A, et al. Leuk Res. 2018 Sep;72:52-58. doi: 10.1016/j.leukres.2018.07.025. Epub 2018 Aug 3. Leuk Res. 2018. PMID: 30098518 Review. - C-terminal binding protein 2 is a novel tumor suppressor targeting the MYC-IRF4 axis in multiple myeloma.
Cheung CHY, Cheng CK, Leung KT, Zhang C, Ho CY, Luo X, Kam AYF, Xia T, Wan TSK, Pitts HA, Chan NPH, Cheung JS, Wong RSM, Zhang XB, Ng MHL. Cheung CHY, et al. Blood Adv. 2024 May 14;8(9):2217-2234. doi: 10.1182/bloodadvances.2023010218. Blood Adv. 2024. PMID: 38457926 Free PMC article. - Impact of hypoxia on the pathogenesis and therapy resistance in multiple myeloma.
Ikeda S, Tagawa H. Ikeda S, et al. Cancer Sci. 2021 Oct;112(10):3995-4004. doi: 10.1111/cas.15087. Epub 2021 Aug 9. Cancer Sci. 2021. PMID: 34310776 Free PMC article. Review.
Cited by
- MMSET stimulates myeloma cell growth through microRNA-mediated modulation of c-MYC.
Min DJ, Ezponda T, Kim MK, Will CM, Martinez-Garcia E, Popovic R, Basrur V, Elenitoba-Johnson KS, Licht JD. Min DJ, et al. Leukemia. 2013 Mar;27(3):686-94. doi: 10.1038/leu.2012.269. Epub 2012 Sep 13. Leukemia. 2013. PMID: 22972034 Free PMC article. - Anticancer activity of the cholesterol exporter ABCA1 gene.
Smith B, Land H. Smith B, et al. Cell Rep. 2012 Sep 27;2(3):580-90. doi: 10.1016/j.celrep.2012.08.011. Epub 2012 Sep 13. Cell Rep. 2012. PMID: 22981231 Free PMC article. - Targeted immunotherapy using anti-CD138-interferon α fusion proteins and bortezomib results in synergistic protection against multiple myeloma.
Vasuthasawat A, Yoo EM, Trinh KR, Lichtenstein A, Timmerman JM, Morrison SL. Vasuthasawat A, et al. MAbs. 2016 Oct;8(7):1386-1397. doi: 10.1080/19420862.2016.1207030. Epub 2016 Jun 30. MAbs. 2016. PMID: 27362935 Free PMC article. - Selective inhibition of tumor oncogenes by disruption of super-enhancers.
Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Lovén J, et al. Cell. 2013 Apr 11;153(2):320-34. doi: 10.1016/j.cell.2013.03.036. Cell. 2013. PMID: 23582323 Free PMC article. - Accelerated development of chronic lymphocytic leukemia in New Zealand Black mice expressing a low level of interferon regulatory factor 4.
Ma S, Shukla V, Fang L, Gould KA, Joshi SS, Lu R. Ma S, et al. J Biol Chem. 2013 Sep 13;288(37):26430-40. doi: 10.1074/jbc.M113.475913. Epub 2013 Jul 29. J Biol Chem. 2013. PMID: 23897826 Free PMC article.
References
- Mittrucker HW, et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–3. - PubMed
- Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–82. - PubMed
- Sciammas R, et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA113992/CA/NCI NIH HHS/United States
- CA97513/CA/NCI NIH HHS/United States
- R33 CA097513/CA/NCI NIH HHS/United States
- Z99 CA999999/ImNIH/Intramural NIH HHS/United States
- R01 CA113992/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases