Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline - PubMed (original) (raw)
. 2008 Jul 11;31(1):79-90.
doi: 10.1016/j.molcel.2008.06.003. Epub 2008 Jun 19.
Marloes P Bagijn, Leonard D Goldstein, Julie R Woolford, Nicolas J Lehrbach, Alexandra Sapetschnig, Heeran R Buhecha, Michael J Gilchrist, Kevin L Howe, Rory Stark, Nik Matthews, Eugene Berezikov, René F Ketting, Simon Tavaré, Eric A Miska
Affiliations
- PMID: 18571451
- PMCID: PMC3353317
- DOI: 10.1016/j.molcel.2008.06.003
Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline
Partha P Das et al. Mol Cell. 2008.
Abstract
The Piwi proteins of the Argonaute superfamily are required for normal germline development in Drosophila, zebrafish, and mice and associate with 24-30 nucleotide RNAs termed piRNAs. We identify a class of 21 nucleotide RNAs, previously named 21U-RNAs, as the piRNAs of C. elegans. Piwi and piRNA expression is restricted to the male and female germline and independent of many proteins in other small-RNA pathways, including DCR-1. We show that Piwi is specifically required to silence Tc3, but not other Tc/mariner DNA transposons. Tc3 excision rates in the germline are increased at least 100-fold in piwi mutants as compared to wild-type. We find no evidence for a Ping-Pong model for piRNA amplification in C. elegans. Instead, we demonstrate that Piwi acts upstream of an endogenous siRNA pathway in Tc3 silencing. These data might suggest a link between piRNA and siRNA function.
Figures
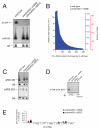
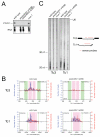
Figure 1. The 21U-RNAs are C. elegans piRNAs
(A) Northern blot showing that 21UR-1 is not detected in RNA (40 _μ_g) isolated from two independent piwi double-mutants: piwi(n4357; n4358) and piwi(n4503; nDf57), whereas miR-52 is expressed in both mutants (same blot re-probed). Antisense DNA probes were used for 21UR-1 and miR-52. A U6 northern blot is shown as loading control. piwi(n4357; n4358) is an abbreviation for prg-1(n4357); prg-2( n4358). piwi(n4503; nDf57) is an abbreviation for prg-1(n4503); prg-2( nDf57). (B) High-throughput sequencing reveals that the expression of many 21U-RNAs is dramatically reduced in piwi mutants. 21U-RNAs cloned from 5′ dependent wild-type and piwi(n4357; n4358) mutant libraries. Frequencies are shown for wild-type (blue) and piwi mutant (red) for the 400 most abundant 21U-RNAs in wild-type, plotted in the order of their wild-type frequency. Read frequencies were obtained by dividing the number of reads for a given 21U-RNA by the total number of reads from the same library (left-hand y-axis). The corresponding absolute number of reads are indicated in the right-hand y-axes. 21U-RNAs for which frequencies are shown include the 21U-RNA with most reads in the piwi mutant library (21UR-3224), which was sequenced 8 times in the piwi mutant and 2,127 times in wild-type. (C) Expression of a 23 nucleotide antisense RNA (siR23-69) and a 26 nucleotide antisense RNA (siR26-263) is not affected in piwi mutants (northern blotting, 40 _μ_g total RNA, antisense DNA probes). (D) Immunoprecipitation followed by RT-PCR for 21UR-5101 reveals that 21U-RNAs are associated with PRG-1 in C. elegans extracts. (E) Quantitative RT-PCR of seven 21U-RNAs demonstrates that Piwi is not essential for 21U-RNA biogenesis. Total RNA was extracted from 12 hour adult C. elegans. Expression levels shown are relative to levels in wild-type RNA. miR-52 expression was used as an internal control. Data are from three independent biological replicates. Error bars represent standard error of the mean.
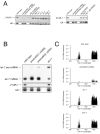
Figure 2. piRNA biogenesis does not require many known small RNA pathway proteins
(A) 21UR-1 northern blotting of total RNA of wild-type and mutant young adult C. elegans. In the case of alg-1; alg-2(RNAi), alg-1 mutant L1 larvae were transferred to alg-2 RNAi feeding plates and young adult animals were harvested. A U6 northern blot is shown as loading control. See Table S1 in Supplemental Data for quantification of these results. (B) 21UR-1 northern blotting of total RNA of wild-type and mutant young adult C. elegans. dcr-1 mutant animals used were homozygous animals derived from heterozygous mothers. To test for loss of DCR-1 activity in dcr-1 mutants, let-7 miRNA and pre-miRNA is shown. A U6 northern blot is shown as loading control. (C) Distribution of 21U-RNAs on chromosome IV as detected by high-throughput sequencing of 5′ dependent wild-type, piwi(n4357; n4358), dcr-1 and mut-7 mutant libraries. Frequencies for a given 21U-RNA and locus were obtained by correcting the number of reads for multiple alignments and dividing by the total number of reads from the same library. Cumulative frequencies were plotted for non-overlapping 100 kb windows along chromosome IV.
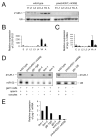
Figure 3. Piwi and piRNAs are restricted to the male and female germline
(A) Profile of 21U-R1 expression during development. E, embryo. L1-L4, larval stages 1-4. YA, 12-hour adult. A, 24-48-hour adult. A U6 northern blot is shown as loading control. (B) Quantitative RT-PCR of 21UR-5101. miR-52 expression was used as an internal control. Data are from three independent biological replicates. Error bars represent standard error of the mean. (C) Quantitative RT-PCR of prg-1 mRNA. Actin mRNA was used as an internal control. Data are from three independent biological replicates. Error bars represent standard error of the mean. (D) Northern blot showing that piRNA expression is restricted to the male and female germline. glp-1(gf), glp-1(lf), glp-4, fem-1 and fem-3 L1 larvae were grown to 12-hour adult stage at 25°C. 20 _μ_g of total RNA was loaded in each lane. U6 and miR-52 northern blots are shown as loading controls. (E) Quantitative RT-PCR of prg-1 mRNA. Actin mRNA was used as an internal control.
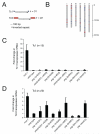
Figure 4. Piwi is required to inhibit Tc3 transposase expression
(A) Diagram of the genomic structure of the two most common DNA transposons in C. elegans, Tc1 and Tc3. Tc1 and Tc3 are flanked by inverted repeats and encode a single spliced transcript for transposase. bp, base pairs. n, number of copies in the wild-type strain N2. (B) Distribution of Tc1 and Tc3 transposons in the C. elegans genome. (C), (D) Quantitative RT-PCR of Tc1 or Tc3 transposase mRNA. As the genomic copies of Tc1 and Tc3 have minor sequence variations, the number of transposon loci amplified by each qRT-PCR primer pair are shown (n). Actin mRNA was used as an internal control. Expression levels shown are relative to levels from wild-type RNA. Data are from three independent biological replicates. Error bars represent standard error of the mean.
Figure 5. Piwi acts upstream of endogenous siRNAs in Tc3 silencing
(A) piRNA expression is independent of MUT-7. 21UR-1 northern blotting of total RNA of wild-type and mutant young adult C. elegans. Total RNA is shown as loading control (GelStar). (B) Tc3 associated small RNAs are absent in piwi mutants. Small RNAs mapping to the loci of Tc3 (top) and Tc1 (bottom) on chromosome I as identified by high-throughput sequencing of 5′ independent wild-type (left) and piwi mutant (right) libraries. Inverted repeat and exon sequences are indicated in green and pink respectively. The number of aligned sequence reads (blue) and number of aligned unique sequences (red) were plotted for each base pair position, with the top and bottom graph in each panel corresponding to the antisense and sense strand relative to the transposase transcript. Read and sequence counts were corrected for multiple alignments to the genome. The total number of reads from wild-type and piwi mutant libraries were comparable (2,963,895 and 3,017,027 of reads with perfect matches to the reference genome respectively). (C) Tc3 transposase antisense siRNAs are dramatically reduced in piwi mutants. RNase protection assay using sense fragments of Tc1 and Tc3 transposase. Sense siRNAs were not detected above background levels using this assay (see Figure S4 in Supplemental Data). U6 was used as an internal control, but its concentration had to be titrated down due to some interference with small RNA detection (Figure S5).
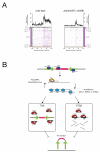
Figure 6. Piwi and piRNAs may act upstream of siRNAs
(A) Small RNAs mapping to the opposite strand of nearby 21U-RNA loci show a preference for locations downstream of the 21U-RNA locus and were reduced in the piwi mutant (right) as compared to wild-type (left). Proximate small RNAs on the same strand as the 21U-RNA were also reduced in the piwi mutant (Figure S7). (Bottom) Rows correspond to 6,021 21U-RNA loci, ordered by genomic position with colors representing different chromosomes. For a given row (21U-RNA locus) dots correspond to the relative position of nearby antisense small RNAs as defined by the distance of the 5′ end of the cloned small RNA relative to the 21U-RNA 5′ end. (Top) Shown is the frequency of distances between 5′ ends of 21U-RNAs and antisense small RNAs. Frequencies were based on the normalized number of loci for 5′ unique sequences. (B) A speculative model of the role of Piwi in Tc3 silencing. TGS, transcriptional gene silencing. PTGS, post-transcriptional gene silencing. See Discussion for an explanation.
Similar articles
- PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans.
Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, Conte D Jr, Luo S, Schroth GP, Carrington JC, Bartel DP, Mello CC. Batista PJ, et al. Mol Cell. 2008 Jul 11;31(1):67-78. doi: 10.1016/j.molcel.2008.06.002. Epub 2008 Jun 19. Mol Cell. 2008. PMID: 18571452 Free PMC article. - C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts.
Lee HC, Gu W, Shirayama M, Youngman E, Conte D Jr, Mello CC. Lee HC, et al. Cell. 2012 Jul 6;150(1):78-87. doi: 10.1016/j.cell.2012.06.016. Epub 2012 Jun 25. Cell. 2012. PMID: 22738724 Free PMC article. - Maternal piRNAs Are Essential for Germline Development following De Novo Establishment of Endo-siRNAs in Caenorhabditis elegans.
de Albuquerque BF, Placentino M, Ketting RF. de Albuquerque BF, et al. Dev Cell. 2015 Aug 24;34(4):448-56. doi: 10.1016/j.devcel.2015.07.010. Epub 2015 Aug 13. Dev Cell. 2015. PMID: 26279485 - Roles of piRNAs in transposon and pseudogene regulation of germline mRNAs and lncRNAs.
Wang C, Lin H. Wang C, et al. Genome Biol. 2021 Jan 8;22(1):27. doi: 10.1186/s13059-020-02221-x. Genome Biol. 2021. PMID: 33419460 Free PMC article. Review. - The piRNA pathway in Drosophila ovarian germ and somatic cells.
Sato K, Siomi MC. Sato K, et al. Proc Jpn Acad Ser B Phys Biol Sci. 2020;96(1):32-42. doi: 10.2183/pjab.96.003. Proc Jpn Acad Ser B Phys Biol Sci. 2020. PMID: 31932527 Free PMC article. Review.
Cited by
- A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi.
Tsai HY, Chen CC, Conte D Jr, Moresco JJ, Chaves DA, Mitani S, Yates JR 3rd, Tsai MD, Mello CC. Tsai HY, et al. Cell. 2015 Jan 29;160(3):407-19. doi: 10.1016/j.cell.2015.01.010. Cell. 2015. PMID: 25635455 Free PMC article. - Identification and characterization of argonaute protein, Ago2 and its associated small RNAs in Schistosoma japonicum.
Cai P, Piao X, Hou N, Liu S, Wang H, Chen Q. Cai P, et al. PLoS Negl Trop Dis. 2012;6(7):e1745. doi: 10.1371/journal.pntd.0001745. Epub 2012 Jul 31. PLoS Negl Trop Dis. 2012. PMID: 22860145 Free PMC article. - RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond.
Castel SE, Martienssen RA. Castel SE, et al. Nat Rev Genet. 2013 Feb;14(2):100-12. doi: 10.1038/nrg3355. Nat Rev Genet. 2013. PMID: 23329111 Free PMC article. Review. - The USTC co-opts an ancient machinery to drive piRNA transcription in C. elegans.
Weng C, Kosalka J, Berkyurek AC, Stempor P, Feng X, Mao H, Zeng C, Li WJ, Yan YH, Dong MQ, Morero NR, Zuliani C, Barabas O, Ahringer J, Guang S, Miska EA. Weng C, et al. Genes Dev. 2019 Jan 1;33(1-2):90-102. doi: 10.1101/gad.319293.118. Epub 2018 Dec 19. Genes Dev. 2019. PMID: 30567997 Free PMC article. - piRNA processing by a trimeric Schlafen-domain nuclease.
Podvalnaya N, Bronkhorst AW, Lichtenberger R, Hellmann S, Nischwitz E, Falk T, Karaulanov E, Butter F, Falk S, Ketting RF. Podvalnaya N, et al. Nature. 2023 Oct;622(7982):402-409. doi: 10.1038/s41586-023-06588-2. Epub 2023 Sep 27. Nature. 2023. PMID: 37758951 Free PMC article.
References
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. - PubMed
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. - PubMed
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials