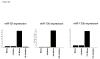miRNA expression in the failing human heart: functional correlates - PubMed (original) (raw)
miRNA expression in the failing human heart: functional correlates
Carmen Sucharov et al. J Mol Cell Cardiol. 2008 Aug.
Abstract
MicroRNAs (miRNAs) are small, noncoding ~22-nucleotide regulatory RNAs that are key regulators of gene expression programs. Their role in the context of the cardiovascular system has only recently begun to be explored; however, changes in the expression of miRNAs have been associated with cardiac development and with several pathophysiological states including myocardial hypertrophy and heart failure. We demonstrate that miRNA expression patterns are distinct in two types of heart failure: idiopathic dilated cardiomyopathy and ischemic cardiomyopathy. To pursue the observation that changes in expression levels of individual miRNAs are functionally relevant, microRNA mimics and inhibitors to miR-92, miR-100 and miR-133b were expressed in primary cultures of neonatal rat cardiac myocytes. These studies demonstrated that over-expression of miR-100 is involved in the beta-adrenergic receptor-mediated repression of "adult" cardiac genes (i.e., alpha-myosin heavy chain, SERCA2a), and that over-expression of miR-133b prevents changes in gene expression patterns mediated by beta-adrenergic receptor stimulation. In conclusion, some miRNA expression patterns appear to be unique to the etiology of cardiomyopathy and changes in the expression level of miRs 100 and 133b contribute to regulation of the fetal gene program. It is likely that this miR-directed reprogramming of key remodeling genes is involved in the establishment and progression of common human cardiomyopathies.
Figures
Figure 1
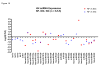
miRNA expression profiles in samples obtained from non-failing (NF), idiopathic cardiomyopathy (IDC) and ischemic (ISC) patients. (A) Relative expression of miRNAs is expressed at the log base 2 ratio of failing (F)/nonfailing (NF). NF vs. IDC (red), NF vs. ISC (blue). Only miRNAs with a p-value < 0.10 as determined by t-Test are shown. (B) Using miRNA specific ABI primers, the relative expression of a subset of miRNAs was confirmed by RT-PCR, as described in Methods.
Figure 1
miRNA expression profiles in samples obtained from non-failing (NF), idiopathic cardiomyopathy (IDC) and ischemic (ISC) patients. (A) Relative expression of miRNAs is expressed at the log base 2 ratio of failing (F)/nonfailing (NF). NF vs. IDC (red), NF vs. ISC (blue). Only miRNAs with a p-value < 0.10 as determined by t-Test are shown. (B) Using miRNA specific ABI primers, the relative expression of a subset of miRNAs was confirmed by RT-PCR, as described in Methods.
Figure 2
Over-expression or down-regulation of a subset of miRNAs in NRVMs regulates β-AR changes in gene expression. (A) Changes in miR-92, miR-133b and miR-100 abundance upon transfection with microRNA mimics or microRNA inhibitors. miRNA abundance was determined by RT-PCR. (B-D) Changes in fetal gene expression in NRVMs transfected with microRNA mimic or microRNA inhibitors. Cells were treated with 10-7M ISO (black bars) 24 hours after transfection and harvested 48 hours after treatment. Gene expression was measured by RT-PCR. Results were normalized to 18S rRNA expression levels and were compared to cells transfected with a control mimic or inhibitor, defined as 1 (line at 1). The graphs represent an average of 3-4 individual experiments.
Figure 2
Over-expression or down-regulation of a subset of miRNAs in NRVMs regulates β-AR changes in gene expression. (A) Changes in miR-92, miR-133b and miR-100 abundance upon transfection with microRNA mimics or microRNA inhibitors. miRNA abundance was determined by RT-PCR. (B-D) Changes in fetal gene expression in NRVMs transfected with microRNA mimic or microRNA inhibitors. Cells were treated with 10-7M ISO (black bars) 24 hours after transfection and harvested 48 hours after treatment. Gene expression was measured by RT-PCR. Results were normalized to 18S rRNA expression levels and were compared to cells transfected with a control mimic or inhibitor, defined as 1 (line at 1). The graphs represent an average of 3-4 individual experiments.
Figure 2
Over-expression or down-regulation of a subset of miRNAs in NRVMs regulates β-AR changes in gene expression. (A) Changes in miR-92, miR-133b and miR-100 abundance upon transfection with microRNA mimics or microRNA inhibitors. miRNA abundance was determined by RT-PCR. (B-D) Changes in fetal gene expression in NRVMs transfected with microRNA mimic or microRNA inhibitors. Cells were treated with 10-7M ISO (black bars) 24 hours after transfection and harvested 48 hours after treatment. Gene expression was measured by RT-PCR. Results were normalized to 18S rRNA expression levels and were compared to cells transfected with a control mimic or inhibitor, defined as 1 (line at 1). The graphs represent an average of 3-4 individual experiments.
Figure 2
Over-expression or down-regulation of a subset of miRNAs in NRVMs regulates β-AR changes in gene expression. (A) Changes in miR-92, miR-133b and miR-100 abundance upon transfection with microRNA mimics or microRNA inhibitors. miRNA abundance was determined by RT-PCR. (B-D) Changes in fetal gene expression in NRVMs transfected with microRNA mimic or microRNA inhibitors. Cells were treated with 10-7M ISO (black bars) 24 hours after transfection and harvested 48 hours after treatment. Gene expression was measured by RT-PCR. Results were normalized to 18S rRNA expression levels and were compared to cells transfected with a control mimic or inhibitor, defined as 1 (line at 1). The graphs represent an average of 3-4 individual experiments.
Figure 3
Over-expression or down-regulation of miRNA-133b in NRVMs regulates cellular hypertrophy. Cells were transfected with miR-133b mimic or inhibitors and cell surface area was quantified. (A) Immunofluorescence with anti-actinin antibody. (B) Cell size was measured using the Image J software. A total of 30 cells from 3 different fields were measured.
Figure 3
Over-expression or down-regulation of miRNA-133b in NRVMs regulates cellular hypertrophy. Cells were transfected with miR-133b mimic or inhibitors and cell surface area was quantified. (A) Immunofluorescence with anti-actinin antibody. (B) Cell size was measured using the Image J software. A total of 30 cells from 3 different fields were measured.
Comment in
- On the road to the definition of the cardiac miRNome in human disease states.
Latronico MV, Condorelli G. Latronico MV, et al. J Mol Cell Cardiol. 2008 Aug;45(2):162-4. doi: 10.1016/j.yjmcc.2008.05.018. Epub 2008 Jun 3. J Mol Cell Cardiol. 2008. PMID: 18619609 No abstract available.
Similar articles
- Inhibition of miR-208b improves cardiac function in titin-based dilated cardiomyopathy.
Zhou Q, Schötterl S, Backes D, Brunner E, Hahn JK, Ionesi E, Aidery P, Sticht C, Labeit S, Kandolf R, Gawaz M, Gramlich M. Zhou Q, et al. Int J Cardiol. 2017 Mar 1;230:634-641. doi: 10.1016/j.ijcard.2016.12.171. Epub 2016 Dec 28. Int J Cardiol. 2017. PMID: 28065693 - MiR-1-3p that correlates with left ventricular function of HCM can serve as a potential target and differentiate HCM from DCM.
Li M, Chen X, Chen L, Chen K, Zhou J, Song J. Li M, et al. J Transl Med. 2018 Jun 9;16(1):161. doi: 10.1186/s12967-018-1534-3. J Transl Med. 2018. PMID: 29885652 Free PMC article. - Identification of micro-RNA networks in end-stage heart failure because of dilated cardiomyopathy.
Zhu X, Wang H, Liu F, Chen L, Luo W, Su P, Li W, Yu L, Yang X, Cai J. Zhu X, et al. J Cell Mol Med. 2013 Sep;17(9):1173-87. doi: 10.1111/jcmm.12096. Epub 2013 Sep 2. J Cell Mol Med. 2013. PMID: 23998897 Free PMC article. - MicroRNAs--regulators of signaling networks in dilated cardiomyopathy.
Naga Prasad SV, Karnik SS. Naga Prasad SV, et al. J Cardiovasc Transl Res. 2010 Jun;3(3):225-34. doi: 10.1007/s12265-010-9177-7. Epub 2010 May 1. J Cardiovasc Transl Res. 2010. PMID: 20560044 Free PMC article. Review. - Role of microRNAs in cardiac hypertrophy and heart failure.
Wang N, Zhou Z, Liao X, Zhang T. Wang N, et al. IUBMB Life. 2009 Jun;61(6):566-71. doi: 10.1002/iub.204. IUBMB Life. 2009. PMID: 19472179 Review.
Cited by
- MicroRNAs in heart failure: Non-coding regulators of metabolic function.
Zhang X, Schulze PC. Zhang X, et al. Biochim Biophys Acta. 2016 Dec;1862(12):2276-2287. doi: 10.1016/j.bbadis.2016.08.009. Epub 2016 Aug 18. Biochim Biophys Acta. 2016. PMID: 27544699 Free PMC article. Review. - Detection and comparison of microRNA expression in the serum of Doberman Pinschers with dilated cardiomyopathy and healthy controls.
Steudemann C, Bauersachs S, Weber K, Wess G. Steudemann C, et al. BMC Vet Res. 2013 Jan 17;9:12. doi: 10.1186/1746-6148-9-12. BMC Vet Res. 2013. PMID: 23327631 Free PMC article. - A cardiac-enriched microRNA, miR-378, blocks cardiac hypertrophy by targeting Ras signaling.
Nagalingam RS, Sundaresan NR, Gupta MP, Geenen DL, Solaro RJ, Gupta M. Nagalingam RS, et al. J Biol Chem. 2013 Apr 19;288(16):11216-32. doi: 10.1074/jbc.M112.442384. Epub 2013 Feb 27. J Biol Chem. 2013. PMID: 23447532 Free PMC article. Retracted. - Pharmacoepigenetics in heart failure.
Mateo Leach I, van der Harst P, de Boer RA. Mateo Leach I, et al. Curr Heart Fail Rep. 2010 Jun;7(2):83-90. doi: 10.1007/s11897-010-0011-y. Curr Heart Fail Rep. 2010. PMID: 20424992 Free PMC article. Review. - MicroRNA profiling and head and neck cancer.
Liu X, Chen Z, Yu J, Xia J, Zhou X. Liu X, et al. Comp Funct Genomics. 2009;2009:837514. doi: 10.1155/2009/837514. Epub 2009 Jun 1. Comp Funct Genomics. 2009. PMID: 19753298 Free PMC article.
References
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001 Dec 28;107(7):823–6. - PubMed
- Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, et al. Altered microRNA expression in human heart disease. Physiol Genomics. 2007 Nov 14;31(3):367–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL048014/HL/NHLBI NIH HHS/United States
- R01 HL051239-15/HL/NHLBI NIH HHS/United States
- R01 HL051239/HL/NHLBI NIH HHS/United States
- HL51239/HL/NHLBI NIH HHS/United States
- HL088708/HL/NHLBI NIH HHS/United States
- K01 HL088708/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources