Characterization of beta-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis - PubMed (original) (raw)
Characterization of beta-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis
Chutikarn Butkinaree et al. J Biol Chem. 2008.
Abstract
Beta-O-linked N-acetylglucosamine is a dynamic post-translational modification involved in protein regulation in a manner similar to phosphorylation. Removal of N-acetylglucosamine is regulated by beta-N-acetylglucosaminidase (O-GlcNAcase), which was previously shown to be a substrate of caspase-3 in vitro. Here we show that O-GlcNAcase is cleaved by caspase-3 into two fragments during apoptosis, an N-terminal fragment containing the O-GlcNAcase active site and a C-terminal fragment containing a region with homology to GCN5 histone acetyl-transferases. The caspase-3 cleavage site of O-GlcNAcase, mapped by Edman sequencing, is a noncanonical recognition site that occurs after Asp-413 of the SVVD sequence in human O-GlcNAcase. A point mutation, D413A, abrogates cleavage by caspase-3 both in vitro and in vivo. Finally, we show that O-GlcNAcase activity is not affected by caspase-3 cleavage because the N- and C-terminal O-GlcNAcase fragments remain associated after the cleavage. Furthermore, when co-expressed simultaneously in the same cell, the N-terminal and C-terminal caspase fragments associate to reconstitute O-GlcNAcase enzymatic activity. These studies support the identification of O-GlcNAcase as a caspase-3 substrate with a novel caspase-3 cleavage site and provide insight about O-GlcNAcase regulation during apoptosis.
Figures
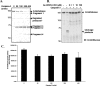
FIGURE 1.
Cleavage of recombinant _O-_GlcNAcase by caspase-3 in vitro. A, 5 μg of human recombinant _O-_GlcNAcase was incubated with indicated amounts of recombinant caspase-3 in caspase-3 assay buffer at 37 °C for 2.5 h. The reaction was stopped by adding Laemmli buffer for Coomassie G-250 staining and Western analysis or 100 μ
m
of caspase-3 inhibitor Ac-DEVD-CHO for _O-_GlcNAcase activity assay. Cleavage mixtures from in vitro caspase-3 cleavage assay were subjected to 7.5% SDS-PAGE followed by Coomassie G-250 staining.B, recombinant _O-_GlcNAcase was incubated with 200 units of caspase-3 in the absence or presence of various concentrations (as indicated) of peptide aldehyde inhibitor Ac-DEVD-CHO and subjected to 7.5% SDS-PAGE followed by immunoblotting (IB) against _O-_GlcNAcase antibody. C, cleavage mixtures from caspase-3 cleavage assay using various amounts of caspase-3 as indicated was analyzed for_O-_GlcNAcase activity in triplicate.
FIGURE 2.
_O-_GlcNAcase is cleaved in cells undergoing apoptosis. A, Jurkat cells were treated with 100 ng/ml of either anti-mouse IgM (control) or anti-Fas CH11 mAb for different time periods. The cell lysates were subjected to 7.5% SDS-PAGE followed by Western blot analysis for_O-_GlcNAcase. Immunoblotting (IB) with PARP and actin antibodies were used as a control for apoptosis and loading respectively._B, O-_GlcNAc levels in Jurkat cells undergoing Fas-mediated apoptosis.C, Jurkat cells were treated with 150 μ
m
H2O2 or PBS (as control) for 8 h. The cell lysates were subjected to Western blot analysis with C-terminally specific_O-_GlcNAcase, PARP, and actin antibodies.
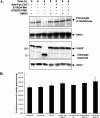
FIGURE 3.
_O-_GlcNAcase is cleaved by caspase-3 during Fas-mediated apoptosis, and caspase-3 cleavage during apoptosis does not affect_O-_GlcNAcase enzymatic activity. A, Jurkat cells were treated with 100 ng/ml of either anti-mouse IgM (control) or anti-Fas CH11 mAb in the presence of dimethyl sulfoxide (DMSO, control) or 50 μ
m
Z-VAD-fmk or 100 μ
m
Z-DEVD-fmk for 6 h, harvested, and subjected to SDS-PAGE, and Western blot analysis for_O-_GlcNAcase cleavage and apoptosis using _O-_GlcNAcase, PARP, and actin antibodies. B, 20 μg of lysate from each sample were analyzed for _O-_GlcNAcase activity in triplicate.
FIGURE 4.
Recombinant _O-_GlcNAcase is cleaved by caspase-3 after Asp-413 of the tetrapeptide sequence SVVD in human _O-_GlcNAcase amino acid sequence. 120 μg of recombinant _O-_GlcNAcase was incubated with 2500 units of caspase-3 at 37 °C for 2.5 h. The cleavage mixtures were subjected to 7.5% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane, which was later stained with Coomassie R-250. Four bands that appeared in the cleavage mixtures but not in the untreated_O-_GlcNAcase were cut out and sequenced in an automated amino acid sequencer (see Fig. 1_A_for the four fragments used in this analysis). A, repetitive yield from N-terminal sequencing indicating the first seven amino acids in the N-terminal sequence of fragment 2. B, schematic view of caspase-3 cleavage of recombinant _O-_GlcNAcase, where the splice variant is also shown (25, 54).Trx_·_tag, thioredoxin tag. C, diagram showing the predicted folded and unfolded regions of human _O-_GlcNAcase using the FoldIndex program (49).D, wild-type _O-_GlcNAcase and D413A mutant were subjected to an in vitro caspase-3 cleavage assay. Cleavage products were analyzed by immunoblotting (IB) with _O-_GlcNAcase antibody.
FIGURE 5.
The point mutation D413A abrogates cleavage of _O-_GlcNAcase during apoptosis in vivo. HeLa cells transfected with empty vector, wild-type _O-_GlcNAcase, or point-mutated D413A_O-_GlcNAcase were treated with 1 μg/ml of either anti-mouse IgM or anti-Fas mAb for 4 h. The cell lysates were subjected to SDS-PAGE and Western blot analysis for _O-_GlcNAcase. IB, immunoblotting.
FIGURE 6.
Both the N and C termini of _O-_GlcNAcase are required for_O-_GlcNAcase activity. HeLa cells were transfected with DNA from empty pRK5 vector, full-length (FL) _O-_GlcNAcase, N-terminal_O-_GlcNAcase (N, amino acids 1–413), C-terminal_O-_GlcNAcase (C, amino acids 414–916), or both N- and C-terminal _O-_GlcNAcase plasmids (N+C). pRK5 vector, full-length _O-_GlcNAcase, and the N-terminal _O-_GlcNAcase plasmids contained c-Myc tag, whereas the C-terminal _O-_GlcNAcase plasmids contained the HA tag. The cells were harvested after 1 day of transfection, extracted, and analyzed for _O-_GlcNAcase expression and_O-_GlcNAcase activity. _A, O-_GlcNAcase activity from transfected HeLa cells. Plasmids and amount of DNA used (for co-transfection of O_-GlcNAcase N and C termini, in μg) are indicated on the_x axis. B and C, cell extracts were subjected to either native PAGE (B) or SDS-PAGE (C) and Western blot analysis for _O-_GlcNAcase using anti-HA and anti-c-Myc antibodies (left and right panels, respectively). Actin detection was used as loading controls. The plasmids and amounts of DNA used were as indicated. IB, immunoblotting.
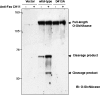
FIGURE 7.
The N and C termini of _O-_GlcNAcase remain associated after caspase-3 cleavage. After a 1-day transfection, HeLa cells were treated with 1 μg/ml of either anti-mouse IgM (control) or anti-Fas CH11 mAb for 4 h. The cell extracts were subjected to immunoprecipitation using antibodies specific to the C terminus of _O-_GlcNAcase. Immunoprecipitated products were subjected to Western blot analysis using antibodies specific to the C-terminal _O-_GlcNAcase (upper panel) and the N-terminal_O-_GlcNAcase (lower panel). 1°, control immunoprecipitation of anti-C-terminal _O-_GlcNAcase antibody in blank extraction buffer. IgY, control immunoprecipitation using anti-chicken IgY antibody instead of anti-C-terminal _O-_GlcNAcase antibody. FL, full length. N+C, N and C termini. IB, immunoblotting.
Similar articles
- Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase.
Wells L, Gao Y, Mahoney JA, Vosseller K, Chen C, Rosen A, Hart GW. Wells L, et al. J Biol Chem. 2002 Jan 18;277(3):1755-61. doi: 10.1074/jbc.m109656200. J Biol Chem. 2002. PMID: 11788610 - Enzymatic characterization of O-GlcNAcase isoforms using a fluorogenic GlcNAc substrate.
Kim EJ, Kang DO, Love DC, Hanover JA. Kim EJ, et al. Carbohydr Res. 2006 Jun 12;341(8):971-82. doi: 10.1016/j.carres.2006.03.004. Epub 2006 Apr 11. Carbohydr Res. 2006. PMID: 16584714 Free PMC article. - O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors.
Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ. Macauley MS, et al. J Biol Chem. 2005 Jul 8;280(27):25313-22. doi: 10.1074/jbc.M413819200. Epub 2005 Mar 28. J Biol Chem. 2005. PMID: 15795231 - Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity.
Copeland RJ, Bullen JW, Hart GW. Copeland RJ, et al. Am J Physiol Endocrinol Metab. 2008 Jul;295(1):E17-28. doi: 10.1152/ajpendo.90281.2008. Epub 2008 Apr 29. Am J Physiol Endocrinol Metab. 2008. PMID: 18445751 Free PMC article. Review. - Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation.
Hu P, Shimoji S, Hart GW. Hu P, et al. FEBS Lett. 2010 Jun 18;584(12):2526-38. doi: 10.1016/j.febslet.2010.04.044. Epub 2010 Apr 22. FEBS Lett. 2010. PMID: 20417205 Review.
Cited by
- _O_-GlcNAcylation: An Emerging Protein Modification Regulating the Hippo Pathway.
Kim E, Kang JG, Jho EH, Yang WH, Cho JW. Kim E, et al. Cancers (Basel). 2022 Jun 18;14(12):3013. doi: 10.3390/cancers14123013. Cancers (Basel). 2022. PMID: 35740678 Free PMC article. Review. - The role of O-GlcNAc signaling in the pathogenesis of diabetic retinopathy.
Semba RD, Huang H, Lutty GA, Van Eyk JE, Hart GW. Semba RD, et al. Proteomics Clin Appl. 2014 Apr;8(3-4):218-31. doi: 10.1002/prca.201300076. Epub 2014 Feb 19. Proteomics Clin Appl. 2014. PMID: 24550151 Free PMC article. Review. - Insights into activity and inhibition from the crystal structure of human O-GlcNAcase.
Elsen NL, Patel SB, Ford RE, Hall DL, Hess F, Kandula H, Kornienko M, Reid J, Selnick H, Shipman JM, Sharma S, Lumb KJ, Soisson SM, Klein DJ. Elsen NL, et al. Nat Chem Biol. 2017 Jun;13(6):613-615. doi: 10.1038/nchembio.2357. Epub 2017 Mar 27. Nat Chem Biol. 2017. PMID: 28346407 - Chemoproteomic profiling of lysine acetyltransferases highlights an expanded landscape of catalytic acetylation.
Montgomery DC, Sorum AW, Meier JL. Montgomery DC, et al. J Am Chem Soc. 2014 Jun 18;136(24):8669-76. doi: 10.1021/ja502372j. Epub 2014 May 30. J Am Chem Soc. 2014. PMID: 24836640 Free PMC article. - Three-dimensional structure of a Streptomyces sviceus GNAT acetyltransferase with similarity to the C-terminal domain of the human GH84 O-GlcNAcase.
He Y, Roth C, Turkenburg JP, Davies GJ. He Y, et al. Acta Crystallogr D Biol Crystallogr. 2014 Jan;70(Pt 1):186-95. doi: 10.1107/S1399004713029155. Epub 2013 Dec 31. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24419391 Free PMC article.
References
- Torres, C. R., and Hart, G. W. (1984) J. Biol. Chem. 2593308 –3317 - PubMed
- Wells, L., Vosseller, K., and Hart, G. W. (2001) Science 2912376 –2378 - PubMed
- Hart, G. W., Housley, M. P., and Slawson, C. (2007) Nature 4461017 –1022 - PubMed
- Whelan, S. A., and Hart, G. W. (2003) Circ. Res. 931047 –1058 - PubMed
- Zachara, N. E., and Hart, G. W. (2002) Chem. Rev. 102431 –438 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials