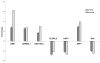Microarray analysis identifies a common set of cellular genes modulated by different HCV replicon clones - PubMed (original) (raw)
Microarray analysis identifies a common set of cellular genes modulated by different HCV replicon clones
Anna Rita Ciccaglione et al. BMC Genomics. 2008.
Abstract
Background: Hepatitis C virus (HCV) RNA synthesis and protein expression affect cell homeostasis by modulation of gene expression. The impact of HCV replication on global cell transcription has not been fully evaluated. Thus, we analysed the expression profiles of different clones of human hepatoma-derived Huh-7 cells carrying a self-replicating HCV RNA which express all viral proteins (HCV replicon system).
Results: First, we compared the expression profile of HCV replicon clone 21-5 with both the Huh-7 parental cells and the 21-5 cured (21-5c) cells. In these latter, the HCV RNA has been eliminated by IFN-alpha treatment. To confirm data, we also analyzed microarray results from both the 21-5 and two other HCV replicon clones, 22-6 and 21-7, compared to the Huh-7 cells. The study was carried out by using the Applied Biosystems (AB) Human Genome Survey Microarray v1.0 which provides 31,700 probes that correspond to 27,868 human genes. Microarray analysis revealed a specific transcriptional program induced by HCV in replicon cells respect to both IFN-alpha-cured and Huh-7 cells. From the original datasets of differentially expressed genes, we selected by Venn diagrams a final list of 38 genes modulated by HCV in all clones. Most of the 38 genes have never been described before and showed high fold-change associated with significant p-value, strongly supporting data reliability. Classification of the 38 genes by Panther System identified functional categories that were significantly enriched in this gene set, such as histones and ribosomal proteins as well as extracellular matrix and intracellular protein traffic. The dataset also included new genes involved in lipid metabolism, extracellular matrix and cytoskeletal network, which may be critical for HCV replication and pathogenesis.
Conclusion: Our data provide a comprehensive analysis of alterations in gene expression induced by HCV replication and reveal modulation of new genes potentially useful for selection of antiviral targets.
Figures
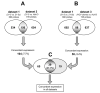
Figure 1
(A) Venn diagram of probes differentially expressed in 21-5 vs. 21-5c (dataset 1) and 21-5 vs. Huh-7 (dataset 2) comparisons. The number of probes differentially expressed in both comparisons was 156, and 104 (7.7%) out of 1344 total probes showed concordant expression (up-regulated or down-regulated in both datasets). (B) Venn diagram of the differentially expressed probes in 21-5 vs. 21-5c (dataset 1) and HCV clones vs. Huh-7 (dataset 3) comparisons. The number of probes differentially expressed in both comparisons was 88, and 58 (4.4%) out of 1327 total probes showed concordant expression. (C) Venn diagram of the 104 and 58 probes identified 39 (31,7%) common probes out of 123.
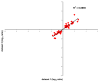
Figure 2
Scatterplot comparison of the log2 ratio of the 58 probes altered in the indicated datasets. Log2 ratios of genes from dataset 3 are plotted on the y-axis and from dataset 1 on the x-axis.
Figure 3
Real-time PCR validation of the microarray data, performed for 7 genes modulated by HCV. Total RNA from the 21-5 and 21-5 cured cell lines was used to assess mRNA levels using real time RT-PCR. Levels were normalized to cellular GAPDH; mRNA levels from 21-5 cured cells were set as the basis for the comparative results. Shown are the means (± SD) of three independent experiments. Fold-changes calculated for the microarray data are also indicated.
Similar articles
- An integrated approach identifies IFN-regulated microRNAs and targeted mRNAs modulated by different HCV replicon clones.
Bruni R, Marcantonio C, Tritarelli E, Tataseo P, Stellacci E, Costantino A, Villano U, Battistini A, Ciccaglione AR. Bruni R, et al. BMC Genomics. 2011 Oct 4;12:485. doi: 10.1186/1471-2164-12-485. BMC Genomics. 2011. PMID: 21970718 Free PMC article. - cDNA microarray analysis to compare HCV subgenomic replicon cells with their cured cells.
Abe K, Ikeda M, Dansako H, Naka K, Shimotohno K, Kato N. Abe K, et al. Virus Res. 2005 Jan;107(1):73-81. doi: 10.1016/j.virusres.2004.06.013. Virus Res. 2005. PMID: 15567036 - Transcriptomic comparison of human hepatoma Huh-7 cell clones with different hepatitis C virus replication efficiencies.
Inoue Y, Murakami K, Hmwe SS, Aizaki H, Suzuki T. Inoue Y, et al. Jpn J Infect Dis. 2007 Jul;60(4):173-8. Jpn J Infect Dis. 2007. PMID: 17642525 - [Replication of hepatitis C virus genome].
Kato N. Kato N. Uirusu. 2008 Dec;58(2):191-8. Uirusu. 2008. PMID: 19374197 Review. Japanese. - [Establishment of cell lines expressing HCV replicon and its applications].
Sakamoto N, Enomoto N. Sakamoto N, et al. Nihon Rinsho. 2004 Jul;62 Suppl 7(Pt 1):116-20. Nihon Rinsho. 2004. PMID: 15359775 Review. Japanese. No abstract available.
Cited by
- Hepatitis C virus (HCV) proteins induce NADPH oxidase 4 expression in a transforming growth factor beta-dependent manner: a new contributor to HCV-induced oxidative stress.
Boudreau HE, Emerson SU, Korzeniowska A, Jendrysik MA, Leto TL. Boudreau HE, et al. J Virol. 2009 Dec;83(24):12934-46. doi: 10.1128/JVI.01059-09. Epub 2009 Oct 7. J Virol. 2009. PMID: 19812163 Free PMC article. - An integrated approach identifies IFN-regulated microRNAs and targeted mRNAs modulated by different HCV replicon clones.
Bruni R, Marcantonio C, Tritarelli E, Tataseo P, Stellacci E, Costantino A, Villano U, Battistini A, Ciccaglione AR. Bruni R, et al. BMC Genomics. 2011 Oct 4;12:485. doi: 10.1186/1471-2164-12-485. BMC Genomics. 2011. PMID: 21970718 Free PMC article. - HCV-Mediated Apoptosis of Hepatocytes in Culture and Viral Pathogenesis.
Silberstein E, Ulitzky L, Lima LA, Cehan N, Teixeira-Carvalho A, Roingeard P, Taylor DR. Silberstein E, et al. PLoS One. 2016 Jun 9;11(6):e0155708. doi: 10.1371/journal.pone.0155708. eCollection 2016. PLoS One. 2016. PMID: 27280444 Free PMC article. - Genome-wide analysis of host mRNA translation during hepatitis C virus infection.
Colman H, Le Berre-Scoul C, Hernandez C, Pierredon S, Bihouée A, Houlgatte R, Vagner S, Rosenberg AR, Féray C. Colman H, et al. J Virol. 2013 Jun;87(12):6668-77. doi: 10.1128/JVI.00538-13. Epub 2013 Apr 3. J Virol. 2013. PMID: 23552407 Free PMC article. - Janus-faced liposomes enhance antimicrobial innate immune response in Mycobacterium tuberculosis infection.
Greco E, Quintiliani G, Santucci MB, Serafino A, Ciccaglione AR, Marcantonio C, Papi M, Maulucci G, Delogu G, Martino A, Goletti D, Sarmati L, Andreoni M, Altieri A, Alma M, Caccamo N, Di Liberto D, De Spirito M, Savage ND, Nisini R, Dieli F, Ottenhoff TH, Fraziano M. Greco E, et al. Proc Natl Acad Sci U S A. 2012 May 22;109(21):E1360-8. doi: 10.1073/pnas.1200484109. Epub 2012 Apr 25. Proc Natl Acad Sci U S A. 2012. PMID: 22538807 Free PMC article.
References
- Ikeda M, Yi M, Li K, Lemon SM. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J Virol. 2002;76:2997–3006. doi: 10.1128/JVI.76.6.2997-3006.2002. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources