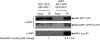Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP - PubMed (original) (raw)
Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP
Qiao Zhao et al. Plant Cell. 2008 Jun.
Abstract
Ran GTPase plays essential roles in multiple cellular processes, including nucleocytoplasmic transport, spindle formation, and postmitotic nuclear envelope (NE) reassembly. The cytoplasmic Ran GTPase activating protein RanGAP is critical to establish a functional RanGTP/RanGDP gradient across the NE and is associated with the outer surface of the NE in metazoan and higher plant cells. Arabidopsis thaliana RanGAP association with the root tip NE requires a family of likely plant-specific nucleoporins combining coiled-coil and transmembrane domains (CC-TMD) and WPP domain-interacting proteins (WIPs). We have now identified, by tandem affinity purification coupled with mass spectrometry, a second family of CC-TMD proteins, structurally similar, yet clearly distinct from the WIP family, that is required for RanGAP NE association in root tip cells. A combination of loss-of-function mutant analysis and protein interaction data indicates that at least one member of each NE-associated CC-TMD protein family is required for RanGAP targeting in root tip cells, while both families are dispensable in other plant tissues. This suggests an unanticipated complexity of RanGAP NE targeting in higher plant cells, contrasting both the single nucleoporin anchor in metazoans and the lack of targeting in fungi and proposes an early evolutionary divergence of the underlying plant and animal mechanisms.
Figures
Figure 1.
Two Novel Coiled-Coil Transmembrane Domain Proteins Interact with Arabidopsis WPP2. (A) Silver-stained gel of tandem affinity purification from TAP-WPP2–expressing 14-d-old Arabidopsis seedlings. Peptides corresponding to WIT1 and WIT2 were identified using tandem mass spectrometry in-gel slices excised from the regions marked with arrowheads. The position of TAP-WPP2 is also marked. (B) Amino acid sequence alignment of WIT1 and WIT2. Identical and similar amino acids are shaded in black and gray, respectively. The four peptides corresponding to the WIT1 protein sequence that were identified by tandem mass spectrometry are boxed in red. The single peptide corresponding to WIT2 protein sequence that was identified in two gel slices by tandem mass spectrometry from TAP-WPP2 pull down is also represented by a red box. Predicted transmembrane domains are marked with a blue box. (C) Domain structures of WIT and WIP protein families are similar. The domain structure is characterized by a presence of extended coiled-coil domain (green) and a single C-terminal transmembrane domain (blue). The position of the transmembrane domain allows classification of both protein families as tail-anchored proteins. WIP protein family members have a bipartite nuclear localization signal motif (red), which has not been identified in the WIT protein sequences.
Figure 2.
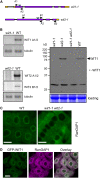
WIT1 Interacts with WPP1, WPP2, RanGAP, and WIP Family Members and with Itself. (A) Interaction of WIT1 with WIP family or WPP domain proteins in yeast two-hybrid assays. RanGAP1ΔC, WPP domain of RanGAP1; RanGAP1ΔN, RanGAP1 without WPP domain; AD, GAL4 activation domain; BD, GAL4 DNA binding domain. Plus (+), positive interaction; minus (−), no interaction. Both WIP1 and WIT1 self-activate as BD fusions, which were therefore not included in this assay. (B) WIT1 interacts with RanGAP1 and 2 and with WIP1 in planta. Interaction with RanGAP1 is abolished by the WPP/AAP mutation in the WPP domain of RanGAP1. FLAG-WIT1 was coexpressed with GFP, RanGAP1-GFP, GFP-RanGAP2, GFP-WIP1, or RanGAP1 (WPP/AAP)-GFP in N. benthamiana. Immunoprecipitation was performed using the anti-GFP antibody, and coimmunoprecipitated protein was detected with the anti-FLAG antibody. (C) WIT1 interacts with WIP proteins and has homodimerization ability in planta. FLAG-WIT1 was coexpressed with GFP, GFP-WIP2a, GFP-WIP3, or GFP-WIT1 in N. benthamiana. Immunoprecipitation was performed using the anti-GFP antibody, and coimmunoprecipitated protein was detected with the anti-FLAG antibody. A nonspecific band detected with the anti-GFP antibody is indicated with an asterisk. (D) WIP1 does not homodimerize in planta. WIP1 was coexpressed with GFP-WIP1 or GFP-WIT1 constructs in N. benthamiana. Immunoprecipitation was performed using the anti-GFP antibody, and coimmunoprecipitated protein was detected with the anti-WIP1 antibody. A nonspecific band detected with the anti-GFP antibody is indicated with an asterisk. (E) WIT1 interacts with endogenous RanGAP1 and WIP1 in Arabidopsis. Samples immunoprecipitated from GFP and GFP-WIT1 transgenic lines using a monoclonal anti-GFP antibody were probed with the anti-RanGAP1 and the anti-WIP1 antibody.
Figure 3.
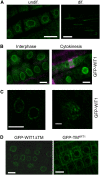
WIT1 Is Targeted to the NE in Arabidopsis Root Tips. (A) Endogenous WIT1 is targeted to the NE in both differentiated cells and undifferentiated cells in Arabidopsis roots. WIT1 is detected by the anti-WIT1 antibody in immunofluorescence analysis. Bars = 10 μm. (B) GFP-WIT1 is targeted to the NE during interphase and cell plate during cytokinesis. Double immunofluorescence of GFP-WIT1 (green) and tubulin (magenta) in Arabidopsis root tip cells. The gain setting of the green channel is increased in the cytokinesis image to highlight the cell plate. Wild-type Arabidopsis plants were used as a negative control, and no fluorescence was detected under the same gain settings (data not shown). Bars = 10 μm. (C) GFP-WIT1 localizes to the NE in a punctate pattern. Confocal images were taken directly from the root tip cells of Arabidopsis expressing GFP-WIT1. The right panel shows three-dimensional maximal projection of confocal images spanning half of the nucleus from Arabidopsis root tip cells. Bar = 10 μm for the left panel and 1 μm for the right panel. (D) The transmembrane domain is dispensable for NE targeting. Root tip cells of a transgenic Arabidopsis line expressing GFP-WIT1 lacking its C-terminal transmembrane domain (GFP-WIT1ΔTM) (left panel) and of a line expressing GFP fused to the WIT1 transmembrane domain alone (GFP-TMWIT1) (right panel). Bars = 10 μm.
Figure 4.
In the wit1-1 wit2-1 Double Mutant, RanGAP1 Is Dislocated from the Root Tip NE. (A) Schematic representation of T-DNA insertions in wit1-1 and wit2-1. Open reading frames, introns, and untranslated regions are indicated by purple boxes, black lines, and yellow boxes, respectively. Light-blue arrowheads are marking T-DNA insertion sites. Positions of primers used for RT-PCR are labeled with black arrowheads. (B) WIT mRNA and protein expression analysis in wit1-1, wit2-1, and wit1-1 wit2-1. The top left panel represents wit1-1 RT-PCR analysis; the bottom left panel represents wit2-1 RT-PCR analysis with two pairs of primers, spanning the T-DNA insertion site (A1/2) or 3′ to the T-DNA insertion site (B1/2). Tubulin, β-tubulin primers used as RT-PCR control. The right panel shows an immunoblot analysis of wit1-1, wit2-1, and wit1-1 wit2-1 using the anti-WIT1 antibody. The position of WIT1 protein is marked with an arrowhead. A section of a Coomassie blue–stained replica gel is shown at the bottom as loading control. (C) Immunofluorescence localization of RanGAP1 in the wild type and wit1-1 wit2-1 double mutant in Arabidopsis root tip cells. Bars = 10 μm. (D) Double immunofluorescence localization of GFP-WIT1 (green) and RanGAP1 (magenta) in the root tip cells of the wit1-1 wit2-1 double mutant transformed with GFP-WIT1. Bar = 10 μm.
Figure 5.
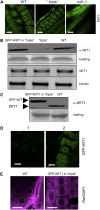
WIT1 Cannot Functionally Replace the WIP Family. (A) Immunofluorescence localization of WIT1 in the wild-type, the wip1-1 wip2-1 wip3-1 triple mutant (“triple”), and the wit1-1 mutant. Bars = 10 μm. (B) WIT1 protein level is reduced in the wip1-1 wip2-1 wip3-1 triple mutant (“triple”). Protein extracts from the wild type, the wip1-1 wip2-1 wip3-1 triple mutant, and GFP-WIP1 in the wip1-1 wip2-1 wip3-1 triple mutant were probed with anti-WIT1 antibody. A section of a Coomassie blue–stained replica gel is shown as loading control. WIT1 mRNA level is not changed in the wip1-1 wip2-1 wip3-1 triple mutant. The third panel from the top shows RT-PCR analysis with WIT1-specific primers. The bottom panel shows tubulin as RT-PCR control. (C) Protein extract from the wild type and GFP-WIT1 in the wip1-1 wip2-1 wip3-1 triple mutant (“triple”) were probed with anti-WIT1 antibody. A Coomassie blue–stained replica gel shown at the bottom represents loading control. (D) Overexpressed GFP-WIT1 is targeted to the NE in the wip1-1 wip2-1 wip3-1 triple mutant. GFP-WIT1 in the wip1-1 wip2-1 wip3-1 triple mutant was detected by live confocal imaging (1) or by immunofluorescence with a polyclonal anti-GFP antibody (ab290) (2). Bars = 10 μm. (E) Immunofluorescence localization of RanGAP1 in root tip cells in the wild-type background (left panel) and in _wip1-1 wip2-1 wip3-1/_GFP-WIT1 (right panel). Bars = 10 μm.
Figure 6.
The Binding Affinity of WIT1 for RanGAP1 Is Increased in the Presence of WIP1. GFP-WIT1 was coexpressed with RanGAP1-GFP in N. benthamiana either in the presence (right two lanes) or absence (left two lanes) of coexpressed WIP1. Immunoprecipitation was performed using the anti-WIT1 antibody, and immunoprecipitated and coimmunoprecipitated proteins were detected with the anti-GFP antibody (top and middle panels) and the anti-WIP1 antibody (bottom panel). The “fold change” of RanGAP1 binding indicated below the lanes was calculated by quantifying the ratio of coimmunoprecipitated RanGAP1-GFP signal to GFP-WIT1 IP signal intensity. The value for GFP-WIT1 binding of RanGAP1-GFP in the absence of WIP1 was set to 1. The “fold change” value is a mean value (±
sd
) obtained from five independent repetitions of the experiment (three biological replicates).
Figure 7.
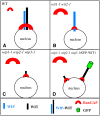
Requirement of Both WIP and WIT Family Members for RanGAP NE Targeting in Root Tip Cells. (A) In the wild type, a complex containing both a WIP and a WIT family member is present at the NE and RanGAP is bound to this complex. (B) In wit1-1 wit2-1, WIP1 abundance and NE association are unchanged. However, in the absence of WIT family members, only a small amount of RanGAP is associated with the NE. (C) In wip1-1 wip2-1 wip3-1, WIT1 abundance is drastically reduced. Remaining WIT1 is likely associated at least in part with the NE but is not sufficient for efficient RanGAP targeting. (D) Overexpressing GFP-WIT1 in wip1-1 wip2-1 wip3-1 does not alter RanGAP localization, indicating that abundant GFP-WIT1 at the NE (which complements wit1-1 wit2-1) is not sufficient for RanGAP targeting in the absence of WIP family members. The size of the symbols in (A) to (D) represents the abundance of the molecule in the respective cellular location.
Similar articles
- Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins.
Xu XM, Meulia T, Meier I. Xu XM, et al. Curr Biol. 2007 Jul 3;17(13):1157-63. doi: 10.1016/j.cub.2007.05.076. Curr Biol. 2007. PMID: 17600715 - Plant-specific mitotic targeting of RanGAP requires a functional WPP domain.
Jeong SY, Rose A, Joseph J, Dasso M, Meier I. Jeong SY, et al. Plant J. 2005 Apr;42(2):270-82. doi: 10.1111/j.1365-313X.2005.02368.x. Plant J. 2005. PMID: 15807788 - A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim.
Rose A, Meier I. Rose A, et al. Proc Natl Acad Sci U S A. 2001 Dec 18;98(26):15377-82. doi: 10.1073/pnas.261459698. Proc Natl Acad Sci U S A. 2001. PMID: 11752475 Free PMC article. - Targeting proteins to the plant nuclear envelope.
Meier I, Zhou X, Brkljacić J, Rose A, Zhao Q, Xu XM. Meier I, et al. Biochem Soc Trans. 2010 Jun;38(3):733-40. doi: 10.1042/BST0380733. Biochem Soc Trans. 2010. PMID: 20491658 Review. - Going green: plants' alternative way to position the Ran gradient.
Meier I, Xu XM, Brkljacic J, Zhao Q, Wang HJ. Meier I, et al. J Microsc. 2008 Aug;231(2):225-33. doi: 10.1111/j.1365-2818.2008.02038.x. J Microsc. 2008. PMID: 18778420 Review.
Cited by
- The central cell nuclear position at the micropylar end is maintained by the balance of F-actin dynamics, but dispensable for karyogamy in Arabidopsis.
Kawashima T, Berger F. Kawashima T, et al. Plant Reprod. 2015 Jun;28(2):103-10. doi: 10.1007/s00497-015-0259-1. Epub 2015 Feb 20. Plant Reprod. 2015. PMID: 25698518 - LINC-complex mediated positioning of the vegetative nucleus is involved in calcium and ROS signaling in Arabidopsis pollen tubes.
Moser M, Kirkpatrick A, Groves NR, Meier I. Moser M, et al. Nucleus. 2020 Dec;11(1):149-163. doi: 10.1080/19491034.2020.1783783. Nucleus. 2020. PMID: 32631106 Free PMC article. - Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination.
Zhou X, Graumann K, Evans DE, Meier I. Zhou X, et al. J Cell Biol. 2012 Jan 23;196(2):203-11. doi: 10.1083/jcb.201108098. J Cell Biol. 2012. PMID: 22270916 Free PMC article. - Interactome of Arabidopsis Thaliana.
Yilmaz M, Paulic M, Seidel T. Yilmaz M, et al. Plants (Basel). 2022 Jan 27;11(3):350. doi: 10.3390/plants11030350. Plants (Basel). 2022. PMID: 35161331 Free PMC article. - RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function.
Tameling WI, Nooijen C, Ludwig N, Boter M, Slootweg E, Goverse A, Shirasu K, Joosten MH. Tameling WI, et al. Plant Cell. 2010 Dec;22(12):4176-94. doi: 10.1105/tpc.110.077461. Epub 2010 Dec 17. Plant Cell. 2010. PMID: 21169509 Free PMC article.
References
- Alber, F., Dodukovskaya, S., Veenhoff, L.M., Zhang, W., Kipper, J., Devos, D., Suprapto, A., Karni-Schmidt, O., Williams, R., Chait, B.T., Rout, M.P., and Sali, A. (2007. b). Determining the architectures of macromolecular assemblies. Nature 450 683–694. - PubMed
- Alber, F., Dodukovskaya, S., Veenhoff, L.M., Zhang, W., Kipper, J., Devos, D., Suprapto, A., Karni-Schmidt, O., Williams, R., Chait, B.T., Sali, A., and Rout, M.P. (2007. a). The molecular architecture of the nuclear pore complex. Nature 450 695–701. - PubMed
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
- Arnaoutov, A., and Dasso, M. (2003). The Ran GTPase regulates kinetochore function. Dev. Cell 5 99–111. - PubMed
- Arnaoutov, A., and Dasso, M. (2005). Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle 4 1161–1165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous