Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly - PubMed (original) (raw)
Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly
Beth A Rasala et al. Mol Biol Cell. 2008 Sep.
Abstract
Assembly of the nuclear pore, gateway to the genome, from its component subunits is a complex process. In higher eukaryotes, nuclear pore assembly begins with the binding of ELYS/MEL-28 to chromatin and recruitment of the large critical Nup107-160 pore subunit. The choreography of steps that follow is largely speculative. Here, we set out to molecularly define early steps in nuclear pore assembly, beginning with chromatin binding. Point mutation analysis indicates that pore assembly is exquisitely sensitive to the change of only two amino acids in the AT-hook motif of ELYS. The dependence on AT-rich chromatin for ELYS binding is borne out by the use of two DNA-binding antibiotics. AT-binding Distamycin A largely blocks nuclear pore assembly, whereas GC-binding Chromomycin A(3) does not. Next, we find that recruitment of vesicles containing the key integral membrane pore proteins POM121 and NDC1 to the forming nucleus is dependent on chromatin-bound ELYS/Nup107-160 complex, whereas recruitment of gp210 vesicles is not. Indeed, we reveal an interaction between the cytoplasmic domain of POM121 and the Nup107-160 complex. Our data thus suggest an order for nuclear pore assembly of 1) AT-rich chromatin sites, 2) ELYS, 3) the Nup107-160 complex, and 4) POM121- and NDC1-containing membrane vesicles and/or sheets, followed by (5) assembly of the bulk of the remaining soluble pore subunits.
Figures
Figure 1.
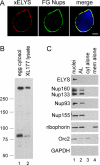
ELYS is abundant in nuclear pores, but not in annulate lamellae (AL) pores. (A) Xenopus reconstituted nuclei were stained with directly labeled anti-xELYS-AF568 (red) and mAb414-AF488 (FG-Nups, green). A merge shows the two localized in an overlapping pattern at the nuclear rim. The DNA is stained with DAPI. Scale bar, 5 μm. (B) Anti-Xenopus ELYS antibody recognizes a full-sized protein band of the expected size of ∼270 kDa and a smaller band (∼80 kDa) in Xenopus egg cytosol (lane 1) and XL177 Xenopus cell lysates (lane 2). (C) Nuclear pores were assembled in the presence of Xenopus egg membranes, cytosol, and sperm chromatin (nuclei, lane 1). AL pores were assembled in the presence of Xenopus egg membranes and cytosol (AL, lane 2). Reactions containing cytosol only (lane 3) or membranes only (lane 4) were used as controls. All reactions were spun through a 30% sucrose cushion, with the heavier components, including the nuclei (lane 1), AL (lane 2), and membrane vesicles (lane 4) pelleting, whereas the soluble proteins (lane 3) remained in the supernatant. The pellets were solubilized with SDS-containing sample buffer, and the presence of ELYS was determined by immunoblot. ELYS was enriched in the nuclear pore assembly reaction, whereas the rest of the soluble Nups tested (Nup160, Nup133, Nup93, and Nup155) purified with both nuclear and AL pores. Orc2, a nuclear protein not associated with pores, is enriched in the purified nuclei (lane 1). Riborphorin, an integral membrane ER protein, copurifies with the membranes. GAPDH, a cytosolic protein not associated with pores, is absent.
Figure 2.
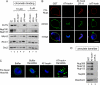
The ELYS C-terminus contains at least two chromatin-binding domains. (A) Cartoons representing the xELYS C-terminal fragments used in this study. The red box represents the AT-hook motif, the black and white striped box represents the arginine (R) to alanine (A) AT-hook motif double point mutant. (B and C) Anchored chromatin-binding assays in which chromatin-coated coverslips were incubated with Xenopus egg cytosol plus the indicated amounts of (B) GST-AT-hook+, GST-AT-hook 2R→A, or (C) GST, GST-AT-hook+, GST-ΔAT-hook. Immunoblots were probed with anti-GST antibody. GST-AT-hook+, GST-AT-hook 2R→A, and GST-ΔAT-hook all bound to the anchored chromatin with varying affinities, whereas GST did not. Dashes represent molecular-weight markers 55 and 40 kDa (B) and 55, 40, 33, and 24 kDa (C).
Figure 3.
The AT-hook motif itself is required for the dominant negative effect of ELYS' C-terminus on nuclear pore assembly. (A) Anchored chromatin binding assay in which chromatin-coated coverslips were incubated with Xenopus egg cytosol plus 10 μM GST, or 5 or 10 μM GST-AT-hook+, GST-AT-hook 2R→A, or GST-ΔAT-hook. Immunoblot analysis using revealed the relative amounts of chromatin binding for endogenous ELYS, Nup107-160 complex members Nup160 and Nup133, Mcm2-7 complex member Mcm3, the RanGEF RCC1, and Orc2. (The apparent mobility shift of RCC1 was not reproducibly observed.) Each lane contains bound protein derived from an equal number of sperm chromatin immobilized on poly-
l
-lysine–treated coverslips, except for the Cyt lane (3 μl of Xenopus egg cytosol diluted 1:10), which is shown as a control for immunoblotting (lane 1). (B) Reconstituted nuclei were assembled in the presence of 15 μM GST, GST-AT-hook+, GST-AT-hook 2R→A, or GST-ΔAT-hook for 1 h. The presence of mature nuclear pores was visualized by staining for the FG-Nups using directly labeled mAb414-AF568 (red). Membrane fusion was determined by continuous DHCC stain of the nuclear membranes (green). DNA was visualized with DAPI (blue, merge). Scale bar, 10 μm. (C) Reconstituted nuclei were assembled in the presence of buffer or GST-AT-hook+, either in the presence or absence of 30 μM RanQ69L-GTP. The FG-Nups, representing mature nuclear pores, were stained with directly labeled mAb414-AF488 (green) and then fixed with 2% formaldehyde before being visualized by fluorescent microscopy. DNA was stained with Hoechst (blue). Scale bar, 10 μm. (D) Annulate lamellae (AL) were assembled by combining purified Xenopus cytosol with membranes in the presence of either 20 μM GST (lane 1) or GST-AT-hook+ (lane 2) for 2 h. The addition of 2 mM GTPγS to reactions containing membranes and cytosol inhibits AL assembly and is shown for comparison (lane 3). All membranes, including membrane-associated proteins, were purified by high-speed centrifugation through a 30% sucrose cushion and were solubilized by SDS-containing sample buffer. Immunoblot analysis using mAb414 revealed that equal amounts of the soluble FG-nucleoporins, Nup358, Nup214, Nup153, and Nup62, assembled into AL supplemented with excess GST or GST-AT-hook+. Integral membrane protein ribophorin served as a loading control.
Figure 4.
The antibiotic Distamycin A inhibits nuclear pore assembly. (A) Reconstituted nuclei were assembled in the presence of buffer, 10 μM Distamycin A (AT-binder), or 10 μM Chromomycin A3 (GC-binder). Nuclear pores were stained with directly labeled mAb414-AF568 (FG-Nups, red). High-magnification views of FG-staining nuclear pores (red) are shown above the red FG-Nup panels. Membrane fusion was determined by continuous DHCC stain of the nuclear membranes (green). DNA was stained with DAPI (blue, merge). Approximately 80% of the nuclei assembled in the presence of 10 μM Distamycin contained little to no visible nuclear pore staining, whereas the majority of the nuclei assembled in the presence of buffer or 10 μM Chromomycin displayed normal nuclear pore staining. Scale bar, 5 μm. (B) Anchored chromatin-binding assay in which chromatin-coated coverslips were incubated with Xenopus egg cytosol plus buffer (lane 2), 10 μM Distamycin A (lane 3), or 10 μM Chromomycin A3 (lane 4). Immunoblot analysis revealed the effect of each antibiotic on the relative amounts of chromatin binding for endogenous ELYS, Nup107-160 complex members Nup160 and Nup133, Mcm2-7 complex member Mcm3, the RanGEF RCC1, and Orc2. Chromatin was not added to the reaction in lane 1 to control for nonspecific binding to the BSA-treated coverslips. In this experiment, each lane, with the exception of lane 1, contains bound protein derived from an equal number of sperm chromatin which had been immobilized on poly-
l
-lysine–treated coverslips. (C) To assess NPC function, nuclei were assembled in control, Distamycin (10 μM), and Chromomycin A3 (10 μM) conditions for 1 h, before incubation with GFP-M9 transport substrate for 5 min. Reactions were stopped with 4% formaldehyde and assessed by fluorescence microscopy for nuclear import. Chromatin was visualized with Hoechst DNA dye. Representative nuclei are shown. (D) Annulate lamellae (AL) were assembled in vitro by incubating Xenopus egg membranes with cytosol, either in the presence of buffer or 10 μM Distamycin A (Dst A; lanes 3 and 4). The purified membrane fractions were probed by immunoblotting (lanes 3–5). A reaction assembled in the presence of 2 mM GTPγS, which blocks membrane fusion and inhibits AL assembly (Meier et al., 1995), was used as a negative control (lane 5). Distamycin A did not affect the assembly of soluble pore proteins Nup160, Nup133, Nup93, or Nup62 into AL. Mem, 3 μl of Xenopus egg membrane fraction diluted 1:20, and cyt, 3 μl of Xenopus egg cytosol diluted 1:10, are shown for comparison (lanes 1 and 2). Ribophorin was used as a loading control.
Figure 5.
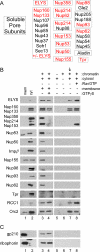
Only ELYS and the Nup107-160 pore subunits bind chromatin in the absence of membranes and RanQ69L-GTP. (A) A diagram representing the known soluble nucleoporin subunits (boxed). It should be noted that we were unable to probe for Aladin because of the lack of an anti-Xenopus Aladin antibody. Nucleoporins highlighted in red were assayed for their ability to bind to anchored chromatin by immunoblotting. (B and C) Anchored chromatin binding assay in which chromatin-coated coverslips were incubated with: Xenopus egg cytosol plus membranes to assemble anchored nuclei (lane 3); egg cytosol plus membranes plus 2 mM GTPγS to block membrane vesicle fusion (lane 4); buffer alone (lane 6), membrane-free Xenopus egg cytosol (lane 7, arrowhead), or the identical cytosol plus 30 μM RanQ69L-GTP (lane 8). A coverslip not coated with chromatin was incubated with cytosol (lane 5) as a control for nonspecific protein binding. In this experiment, Orc2 serves as a loading control. Mem, Xenopus egg membrane fraction diluted 1:20, and cyt, Xenopus egg cytosol diluted 1:10, are shown as a control for immunoblotting with the various antibodies (lanes 1 and 2). It should be noted that we did not detect chromatin binding of Nup358 in the presence of excess RanGTP, as was previously published (Walther et al., 2003b). (C) Immunoblots using antibodies to the integral membrane proteins gp210 and ribophorin show that membrane vesicles are not present in the Xenopus egg cytosol (lanes 2 and 5–8).
Figure 6.
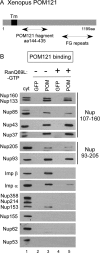
POM121 binds the Nup107-160 and the Nup93-205 complexes in the absence of FG Nups, Nup53, and Nup155. (A) A map of POM121 and the fragment used. (B) The Xenopus POM121 fragment aa 144-435 (POM) was coupled to beads and mixed with Xenopus egg cytosol in pulldown reactions, in the presence or the absence of 10 μM RanQ69L-GTP. his-GFP beads were used as a control. The bound proteins were probed by immunoblotting with different anti-nucleoporin and import factor antibodies, as shown in the figure. Nup160, Nup133, Nup85, Nup43, and Nup37 of the Nup107-160 complex, and Nup93 and Nup205 of the Nup93-205 complex specifically bound to the POM121 fragment. Cyt, Xenopus egg cytosol diluted 1:10, is shown as a control for immunoblotting with the various antibodies (lane 1).
Figure 7.
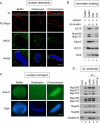
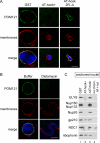
Chromatin-bound ELYS/Nup107-160 complex recruit POM121- and NDC1-containing membrane vesicles. (A) Reconstituted nuclei were assembled in the presence of 15 μM GST, GST-AT-hook+, or GST-AT-hook-2R→A. The presence of POM121 in the nuclear membranes was probed for with directly labeled anti-POM121-AF488 (green). Membranes were stained with R18 membrane dye (red). DNA was stained with Hoechst (blue). Nuclei were fixed before mounting onto slides. Scale bar, 10 μm. (B) Reconstituted nuclei were assembled in the presence of buffer or 10 μm Distamycin A and processed as in A. Scale bar, 10 μm. (C) Anchored nuclei were assembled by incubating chromatin-coated coverslips with membranes and cytosol for 1 h, in the presence of either 15 μM GST, GST-AT-hook+, or GST-ΔAT-hook. A coverslip not treated with chromatin, but incubated with membranes, M, and cytosol, cyt, was used as a control for nonspecific binding (lane 4). The recruitment of integral membrane pore proteins NDC1 and gp210, in the presence (GST and GST-ΔAT-hook, lane 1 and 3) or absence (GST-AT-hook+, lane 2) of chromatin-bound ELYS/Nup107-160 complex, was determined by immunoblot analysis.
Figure 8.
A model for the early steps in NPC assembly. On the basis of our findings, we propose a model for the early steps in NPC assembly. The first step in pore assembly is the binding of ELYS (yellow) to AT-rich chromatin sites, selected via the high-affinity AT-hook motif (black box) and strengthened by its second chromatin binding domain. ELYS then acts as a bridge between the chromatin and the Nup107-160 complex (green; Rasala et al., 2006; Franz et al., 2007; Gillespie et al., 2007). Chromatin-bound ELYS/Nup107-160 complex actively recruits the integral membrane pore proteins POM121 (red) and NDC1 (blue), likely through interaction with the cytosolic domain of POM121. After membrane vesicle fusion, the rest of the soluble pore subunits assemble and a complete nuclear pore is formed. (Note, other vesicles/sheets bind to the chromatin via different linkages such as lamins to form the bulk of the nuclear membranes, but are not shown here.)
Comment in
- Mol Biol Cell. 19:3615.
Similar articles
- Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly.
Lau CK, Delmar VA, Chan RC, Phung Q, Bernis C, Fichtman B, Rasala BA, Forbes DJ. Lau CK, et al. Mol Biol Cell. 2009 Sep;20(18):4043-58. doi: 10.1091/mbc.e09-02-0152. Epub 2009 Jul 29. Mol Biol Cell. 2009. PMID: 19641022 Free PMC article. - ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division.
Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. Rasala BA, et al. Proc Natl Acad Sci U S A. 2006 Nov 21;103(47):17801-6. doi: 10.1073/pnas.0608484103. Epub 2006 Nov 10. Proc Natl Acad Sci U S A. 2006. PMID: 17098863 Free PMC article. - Structural and biochemical analyses of the nuclear pore complex component ELYS identify residues responsible for nucleosome binding.
Kobayashi W, Takizawa Y, Aihara M, Negishi L, Ishii H, Kurumizaka H. Kobayashi W, et al. Commun Biol. 2019 May 3;2:163. doi: 10.1038/s42003-019-0385-7. eCollection 2019. Commun Biol. 2019. PMID: 31069272 Free PMC article. - The Role of Nucleoporin Elys in Nuclear Pore Complex Assembly and Regulation of Genome Architecture.
Shevelyov YY. Shevelyov YY. Int J Mol Sci. 2020 Dec 13;21(24):9475. doi: 10.3390/ijms21249475. Int J Mol Sci. 2020. PMID: 33322130 Free PMC article. Review. - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review.
Cited by
- Identification of Conserved MEL-28/ELYS Domains with Essential Roles in Nuclear Assembly and Chromosome Segregation.
Gómez-Saldivar G, Fernandez A, Hirano Y, Mauro M, Lai A, Ayuso C, Haraguchi T, Hiraoka Y, Piano F, Askjaer P. Gómez-Saldivar G, et al. PLoS Genet. 2016 Jun 24;12(6):e1006131. doi: 10.1371/journal.pgen.1006131. eCollection 2016 Jun. PLoS Genet. 2016. PMID: 27341616 Free PMC article. - The nucleoporin ALADIN regulates Aurora A localization to ensure robust mitotic spindle formation.
Carvalhal S, Ribeiro SA, Arocena M, Kasciukovic T, Temme A, Koehler K, Huebner A, Griffis ER. Carvalhal S, et al. Mol Biol Cell. 2015 Oct 1;26(19):3424-38. doi: 10.1091/mbc.E15-02-0113. Epub 2015 Aug 5. Mol Biol Cell. 2015. PMID: 26246606 Free PMC article. - Structural and functional studies of the 252 kDa nucleoporin ELYS reveal distinct roles for its three tethered domains.
Bilokapic S, Schwartz TU. Bilokapic S, et al. Structure. 2013 Apr 2;21(4):572-80. doi: 10.1016/j.str.2013.02.006. Epub 2013 Mar 14. Structure. 2013. PMID: 23499022 Free PMC article. - Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane.
Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW. Mitchell JM, et al. J Cell Biol. 2010 Nov 1;191(3):505-21. doi: 10.1083/jcb.201007098. Epub 2010 Oct 25. J Cell Biol. 2010. PMID: 20974814 Free PMC article. - Cytosol-dependent membrane fusion in ER, nuclear envelope and nuclear pore assembly: biological implications.
Rafikova ER, Melikov K, Chernomordik LV. Rafikova ER, et al. Nucleus. 2010 Nov-Dec;1(6):487-91. doi: 10.4161/nucl.1.6.13514. Epub 2010 Sep 3. Nucleus. 2010. PMID: 21327091 Free PMC article.
References
- Alber F., et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. - PubMed
- Anand A., Chada K. In vivo modulation of Hmgic reduces obesity. Nat. Genet. 2000;24:377–380. - PubMed
- Anderson D. J., Hetzer M. W. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat. Cell Biol. 2007;9:1160–1166. - PubMed
- Antonin W., Ellenberg J., Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. - PubMed
- Antonin W., Franz C., Haselmann U., Antony C., Mattaj I. W. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol. Cell. 2005;17:83–92. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous