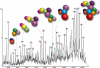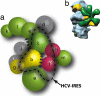Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3 - PubMed (original) (raw)
. 2008 Nov 25;105(47):18139-44.
doi: 10.1073/pnas.0801313105. Epub 2008 Jul 1.
Alan M Sandercock, Christopher S Fraser, Gabriela Ridlova, Elaine Stephens, Matthew R Schenauer, Theresa Yokoi-Fong, Daniel Barsky, Julie A Leary, John W Hershey, Jennifer A Doudna, Carol V Robinson
Affiliations
- PMID: 18599441
- PMCID: PMC2587604
- DOI: 10.1073/pnas.0801313105
Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3
Min Zhou et al. Proc Natl Acad Sci U S A. 2008.
Abstract
The eukaryotic initiation factor 3 (eIF3) plays an important role in translation initiation, acting as a docking site for several eIFs that assemble on the 40S ribosomal subunit. Here, we use mass spectrometry to probe the subunit interactions within the human eIF3 complex. Our results show that the 13-subunit complex can be maintained intact in the gas phase, enabling us to establish unambiguously its stoichiometry and its overall subunit architecture via tandem mass spectrometry and solution disruption experiments. Dissociation takes place as a function of ionic strength to form three stable modules eIF3(c:d:e:l:k), eIF3(f:h:m), and eIF3(a:b:i:g). These modules are linked by interactions between subunits eIF3b:c and eIF3c:h. We confirmed our interaction map with the homologous yeast eIF3 complex that contains the five core subunits found in the human eIF3 and supplemented our data with results from immunoprecipitation. These results, together with the 27 subcomplexes identified with increasing ionic strength, enable us to define a comprehensive interaction map for this 800-kDa species. Our interaction map allows comparison of free eIF3 with that bound to the hepatitis C virus internal ribosome entry site (HCV-IRES) RNA. We also compare our eIF3 interaction map with related complexes, containing evolutionarily conserved protein domains, and reveal the location of subunits containing RNA recognition motifs proximal to the decoding center of the 40S subunit of the ribosome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
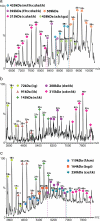
MS of the intact eIF3 revealing a series of well resolved charge states consistent with the predominant species being the intact 13 subunit complex. Masses of the 13 subunits were confirmed in a separate proteomics analysis. The y axis is the relative intensity of the peaks. The inventory of subunits is shown with the radius of each subunit scaled according to mass. (Inset) High-energy MS spectrum of eIF3 from 150 mM AmAc solution, showing a second charge state series (blue star) resulting from the dissociation of eIF3j from the intact complex (purple star).
Fig. 2.
Mass spectra recorded after increasing the ionic strength from 250 mM AmAc (a) to 350 mM (b) and 500 mM (c). Series of subcomplexes, observed with decreasing m/z values as the ionic strength is increased, are assigned based on their masses, the observation of common subunit losses, and from tandem MS.
Fig. 3.
MS spectrum of the yeast eIF3 isolated by tagging subunit eIF3b. Charge state series are assigned on the basis of masses to subcomplexes eIF3i:g, eIF3b:g:i, and eIF3a:b. eIF3i is observed dissociating from the yeast complex at ≈m/z 3,000. (Inset) The interaction network for yeast eIF3 derived from seven subcomplexes observed by MS.
Fig. 4.
Model of the human eIF3 derived from 27 subcomplexes, IP data, and interactions identified in the yeast complex. The complex dissociates into distinct modules in response to changes in ionic strength; in this case, the spectrum shown was recorded at intermediate ionic strength (350 mM AmAc). Arrows denote additional interactions not readily represented in this model.
Fig. 5.
Proposed model for the eIF3:HCV IRES interaction. (a) Subunit organization colored according to signature domains contained within the various subunits. PCI-containing domains (green), MPN domains (red), and RNA recognition motifs (yellow) show direct interactions with the exception of eIF3m and eIF3a. Subunits with no common signature domains are shown in gray. Subunits that are affected by binding of HCV IRES are within the dashed line. The location of subunits satisfies the interaction network and is not indicative of their location within the EM density. (b) EM model of eIF3-IRES-40S complex showing the binding of IRES (yellow) to both eIF3 (green) and the 40S (blue).
Similar articles
- Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit.
Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. Hashem Y, et al. Nature. 2013 Nov 28;503(7477):539-43. doi: 10.1038/nature12658. Epub 2013 Nov 3. Nature. 2013. PMID: 24185006 Free PMC article. - Distinct regions of human eIF3 are sufficient for binding to the HCV IRES and the 40S ribosomal subunit.
Cai Q, Todorovic A, Andaya A, Gao J, Leary JA, Cate JH. Cai Q, et al. J Mol Biol. 2010 Oct 22;403(2):185-96. doi: 10.1016/j.jmb.2010.07.054. Epub 2010 Sep 15. J Mol Biol. 2010. PMID: 20816988 Free PMC article. - Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3).
Sun C, Todorovic A, Querol-Audí J, Bai Y, Villa N, Snyder M, Ashchyan J, Lewis CS, Hartland A, Gradia S, Fraser CS, Doudna JA, Nogales E, Cate JH. Sun C, et al. Proc Natl Acad Sci U S A. 2011 Dec 20;108(51):20473-8. doi: 10.1073/pnas.1116821108. Epub 2011 Dec 1. Proc Natl Acad Sci U S A. 2011. PMID: 22135459 Free PMC article. - eIF3: a versatile scaffold for translation initiation complexes.
Hinnebusch AG. Hinnebusch AG. Trends Biochem Sci. 2006 Oct;31(10):553-62. doi: 10.1016/j.tibs.2006.08.005. Epub 2006 Aug 22. Trends Biochem Sci. 2006. PMID: 16920360 Review. - Hepatitis C Virus Translation Regulation.
Niepmann M, Gerresheim GK. Niepmann M, et al. Int J Mol Sci. 2020 Mar 27;21(7):2328. doi: 10.3390/ijms21072328. Int J Mol Sci. 2020. PMID: 32230899 Free PMC article. Review.
Cited by
- Knockdown of eukaryotic translation initiation factors 3B (EIF3B) inhibits proliferation and promotes apoptosis in glioblastoma cells.
Liang H, Ding X, Zhou C, Zhang Y, Xu M, Zhang C, Xu L. Liang H, et al. Neurol Sci. 2012 Oct;33(5):1057-62. doi: 10.1007/s10072-011-0894-8. Epub 2012 Jan 11. Neurol Sci. 2012. PMID: 22234522 - Structure from Splatter.
Leary JA. Leary JA. ACS Cent Sci. 2015 Dec 23;1(9):475-6. doi: 10.1021/acscentsci.5b00390. Epub 2015 Dec 15. ACS Cent Sci. 2015. PMID: 27163012 Free PMC article. No abstract available. - Two decades of studying non-covalent biomolecular assemblies by means of electrospray ionization mass spectrometry.
Hilton GR, Benesch JL. Hilton GR, et al. J R Soc Interface. 2012 May 7;9(70):801-16. doi: 10.1098/rsif.2011.0823. Epub 2012 Feb 7. J R Soc Interface. 2012. PMID: 22319100 Free PMC article. Review. - An Ion Mobility-Mass Spectrometry Investigation of Monocyte Chemoattractant Protein-1.
Schenauer MR, Leary JA. Schenauer MR, et al. Int J Mass Spectrom. 2009 Oct 15;287(1-3):70-76. doi: 10.1016/j.ijms.2009.02.023. Int J Mass Spectrom. 2009. PMID: 20160907 Free PMC article. - Merging molecular electron microscopy and mass spectrometry by carbon film-assisted endoproteinase digestion.
Richter FM, Sander B, Golas MM, Stark H, Urlaub H. Richter FM, et al. Mol Cell Proteomics. 2010 Aug;9(8):1729-41. doi: 10.1074/mcp.M110.001446. Epub 2010 Jun 8. Mol Cell Proteomics. 2010. PMID: 20530635 Free PMC article.
References
- Prichard PM, Gilbert JM, Shafritz DA, Anderson WF. Factors for the initiation of haemoglobin synthesis by rabbit reticulocyte ribosomes. Nature. 1970;226:511–514. - PubMed
- Valasek L, Hasek J, Nielsen KH, Hinnebusch AG. Dual function of eIF3j/Hcr1p in processing 20 S pre-rRNA and translation initiation. J Biol Chem. 2001;276:43351–43360. - PubMed
- Pestova TV, Lorsch JR, Hellen CUT. The Mechanism of Translation Initiation in Eukaryotes. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases