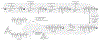Synthesis and biophysical characterization of stabilized alpha-helices of BCL-2 domains - PubMed (original) (raw)
Synthesis and biophysical characterization of stabilized alpha-helices of BCL-2 domains
Gregory H Bird et al. Methods Enzymol. 2008.
Abstract
Rational design of compounds to mimic the functional domains of BCL-2 family proteins requires chemical reproduction of the biologic complexity afforded by the relatively large and folded surfaces of BCL-2 homology (BH) domain peptide alpha-helices. Because the intermolecular handshakes of BCL-2 proteins are so critical to controlling cellular fate, we undertook the development of a toolbox of peptidic ligands that harness the natural potency and specificity of BH alpha-helices to interrogate and potentially medicate the deregulated apoptotic pathways of human disease. To overcome the classic deficiencies of peptide reagents, including loss of bioactive structure in solution, rapid proteolytic degradation in vivo, and cellular impermeability, we developed a new class of compounds based on hydrocarbon stapling of BH3 death domain peptides. Here we describe the chemical synthesis of Stabilized Alpha-Helices of BCL-2 domains or SAHBs, and the analytical methods used to characterize their secondary structure, proteolytic stability, and cellular penetrance.
Figures
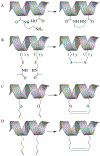
Figure 22.1
Approaches to covalent α-helical stabilization have included the use of (A) lactam bridges, (B) disulfide bridges, (C) ruthenium-catalyzed ring closing metathesis (RCM) of O-allyl serine residues, and (D) RCM of α,α-disubstituted non-natural amino acids bearing alkyl tethers.
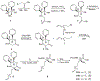
Figure 22.2
Synthetic scheme for generating the chiral α,α-disubstituted non-natural amino acids used to staple bioactive peptides.
Figure 22.3
Design considerations for installing hydrocarbon staples derive from structural data (e.g., BAK BH3/BCL-XL ΔC; PDB 1BXL [Sattler et al., 1997]) that highlight key interacting surfaces to be avoided. Several (i, i + 4) residues indicated as red balls on the noninteracting surface of the BAK BH3 helix are amenable to replacement with the S5 non-natural amino acid.
Figure 22.4
Synthetic scheme for the generation of SAHBs by Fmoc-based solid-phase peptide synthesis and ruthenium-catalyzed olefin metathesis.
Figure 22.5
Circular dichroism spectra demonstrate the increased α-helicity of SAHBs compared to the corresponding unmodified BH3 peptides (Walensky et al., 2006).
Figure 22.6
SAHBs exhibit marked protease resistance compared with unmodified BH3 peptides as assessed by (A) in vitro trypsin degradation assay, (B) ex vivo, and (C) in vivo serum stability assays (Walensky et al., 2004).
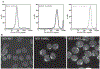
Figure 22.7
Cellular uptake of FITC-BID SAHB and a point mutant derivative, but not the unmodified FITC-BID BH3 peptide, is readily demonstrated by (A) FACS analysis and (B) confocal microscopy of FITC-peptide treated cells in culture. The cellular fluorescence of FITC-SAHB treated cells, as reflected by a shift of the FACS profile to the right compared to FITC-BID BH3–treated cells, corresponds to the intracellular cytosolic localization of FITC-BID SAHB observed by confocal microscopy. (Walensky et al., 2004)
Similar articles
- Dissection of the BCL-2 family signaling network with stabilized alpha-helices of BCL-2 domains.
Pitter K, Bernal F, Labelle J, Walensky LD. Pitter K, et al. Methods Enzymol. 2008;446:387-408. doi: 10.1016/S0076-6879(08)01623-6. Methods Enzymol. 2008. PMID: 18603135 Free PMC article. - Photoreactive stapled peptides to identify and characterize BCL-2 family interaction sites by mass spectrometry.
Lee S, Braun CR, Bird GH, Walensky LD. Lee S, et al. Methods Enzymol. 2014;544:25-48. doi: 10.1016/B978-0-12-417158-9.00002-9. Methods Enzymol. 2014. PMID: 24974285 - Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix.
Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Walensky LD, et al. Science. 2004 Sep 3;305(5689):1466-70. doi: 10.1126/science.1099191. Science. 2004. PMID: 15353804 Free PMC article. - Structural biology of the Bcl-2 family of proteins.
Petros AM, Olejniczak ET, Fesik SW. Petros AM, et al. Biochim Biophys Acta. 2004 Mar 1;1644(2-3):83-94. doi: 10.1016/j.bbamcr.2003.08.012. Biochim Biophys Acta. 2004. PMID: 14996493 Review. - BH3 domains: intracellular death-ligands critical for initiating apoptosis.
Chittenden T. Chittenden T. Cancer Cell. 2002 Sep;2(3):165-6. doi: 10.1016/s1535-6108(02)00128-9. Cancer Cell. 2002. PMID: 12242145 Review.
Cited by
- Found in Translation: How Preclinical Research Is Guiding the Clinical Development of the BCL2-Selective Inhibitor Venetoclax.
Leverson JD, Sampath D, Souers AJ, Rosenberg SH, Fairbrother WJ, Amiot M, Konopleva M, Letai A. Leverson JD, et al. Cancer Discov. 2017 Dec;7(12):1376-1393. doi: 10.1158/2159-8290.CD-17-0797. Epub 2017 Nov 16. Cancer Discov. 2017. PMID: 29146569 Free PMC article. Review. - The Role of Structural Flexibility in Hydrocarbon-Stapled Peptides Designed to Block Viral Infection via Human ACE2 Mimicry.
Jiang S, Tian Y, Nicolaescu V, Mansurov A, Randall G, Tirrell MV, LaBelle JL. Jiang S, et al. Pept Sci (Hoboken). 2024 Nov;116(6):e24375. doi: 10.1002/pep2.24375. Epub 2024 Jul 22. Pept Sci (Hoboken). 2024. PMID: 39866902 - Inhibition of Pro-apoptotic BAX by a noncanonical interaction mechanism.
Barclay LA, Wales TE, Garner TP, Wachter F, Lee S, Guerra RM, Stewart ML, Braun CR, Bird GH, Gavathiotis E, Engen JR, Walensky LD. Barclay LA, et al. Mol Cell. 2015 Mar 5;57(5):873-886. doi: 10.1016/j.molcel.2015.01.014. Epub 2015 Feb 12. Mol Cell. 2015. PMID: 25684204 Free PMC article. - Targeting a helix-in-groove interaction between E1 and E2 blocks ubiquitin transfer.
Cathcart AM, Bird GH, Wales TE, Herce HD, Harvey EP, Hauseman ZJ, Newman CE, Adhikary U, Prew MS, Oo T, Lee S, Engen JR, Walensky LD. Cathcart AM, et al. Nat Chem Biol. 2020 Nov;16(11):1218-1226. doi: 10.1038/s41589-020-0625-7. Epub 2020 Aug 17. Nat Chem Biol. 2020. PMID: 32807965 Free PMC article. - Chemical synthesis of hydrocarbon-stapled peptides for protein interaction research and therapeutic targeting.
Bird GH, Crannell WC, Walensky LD. Bird GH, et al. Curr Protoc Chem Biol. 2011 Sep 1;3(3):99-117. doi: 10.1002/9780470559277.ch110042. Curr Protoc Chem Biol. 2011. PMID: 23801563 Free PMC article.
References
- Albericio F (2004). Developments in peptide and amide synthesis. Curr. Opin. Chem. Bio 8, 211–221. - PubMed
- Banerjee R, Basu G, Chene P, and Roy S (2002). Aib-based peptide backbone as scaffolds for helical peptide mimics. J. Pept. Res 60, 88–94. - PubMed
- Blackwell HE, and Grubbs RH (1994). Highly Efficient Synthesis of Covalently Cross-Linked Peptide Helices by Ring-Closing Metathesis. Angew Chem. Int. Ed 37, 3281–3284. - PubMed
- Bohm G, Muhr R, and Jaenicke R (1992). Quantitative-Analysis of Protein Far Uv Circular-Dichroism Spectra by Neural Networks. Protein Eng. 5, 191–195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5R01CA50239/CA/NCI NIH HHS/United States
- 5P01CA92625/CA/NCI NIH HHS/United States
- P01 CA092625/CA/NCI NIH HHS/United States
- K08HL074049/HL/NHLBI NIH HHS/United States
- K08 HL074049/HL/NHLBI NIH HHS/United States
- K22 CA128886/CA/NCI NIH HHS/United States
- R01 CA050239/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources