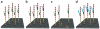Alta-Cyclic: a self-optimizing base caller for next-generation sequencing - PubMed (original) (raw)
Alta-Cyclic: a self-optimizing base caller for next-generation sequencing
Yaniv Erlich et al. Nat Methods. 2008 Aug.
Abstract
Next-generation sequencing is limited to short read lengths and by high error rates. We systematically analyzed sources of noise in the Illumina Genome Analyzer that contribute to these high error rates and developed a base caller, Alta-Cyclic, that uses machine learning to compensate for noise factors. Alta-Cyclic substantially improved the number of accurate reads for sequencing runs up to 78 bases and reduced systematic biases, facilitating confident identification of sequence variants.
Figures
Figure 1
Schematic representation of main Illumina noise factors. (a–d)A DNA cluster comprises identical DNA templates (colored boxes) that are attached to the flow cell. Nascent strands (black boxes) and DNA polymerase (black ovals) are depicted. In the ideal situation, after several cycles the signal (green arrows) is strong, coherent and corresponds to the interrogated position (a). Phasing noise introduces lagging (blue arrows) and leading (red arrow) nascent strands, which transmit a mixture of signals (b). Fading is attributed to loss of material that reduces the signal intensity (c). Changes in the fluorophore cross-talk cause misinterpretation of the received signal (teal arrows; d). For simplicity, the noise factors are presented separately from each other.
Figure 2
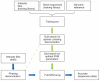
Alta-Cyclic base caller data flow. The training process (green arrows) starts with creation of the training set, beginning with sequences generated by the standard Illumina pipeline, by linking intensity reads and a corresponding genome sequence (the ‘correct’ sequence). Then, two grid searches are used to optimize the parameters to call the bases. After optimization, a final SVM array is created, each of which corresponds to a cycle. In the base-calling stage (blue arrows), the intensity files of the desired library undergo deconvolution to correct for phasing noise using the optimized values and are sent for classification with the SVM array. The output is processed, and sequences and quality scores are reported.
Figure 3
Comparison between Alta-Cyclic and Illumina base caller on the GAII platform. (a) Analysis of the HepG2 RNA library using Alta-Cyclic. The absolute number of additional fully correct reads (in addition to those generated by the Illumina base caller) is indicated by the red line; the fold change of the improvement is indicated by the blue bars. (b) A comparison of fully correct reads for the Tetrahymena micronuclear library by the Illumina base caller and Alta-Cyclic. (c) The average error rate in calls of the artificial SNP locations in the phi X library as a function of the cycle in which they were called. The dashed line represents 1% error rate (Q20). The plot on the right shows the last 18 cycles in a different scale. (d) A comparison of fully correct reads for the phi X library with 1% artificial SNPs. (e) Phi X sequences generated by Alta-Cyclic or Illumina were exhaustively aligned to the reference genome (allowing up to 53 mismatches out of 78). The distribution of alignment scores is shown beginning with an identical number of raw reads for input into each base caller.
Similar articles
- Improved base calling for the Illumina Genome Analyzer using machine learning strategies.
Kircher M, Stenzel U, Kelso J. Kircher M, et al. Genome Biol. 2009;10(8):R83. doi: 10.1186/gb-2009-10-8-r83. Epub 2009 Aug 14. Genome Biol. 2009. PMID: 19682367 Free PMC article. - GemSIM: general, error-model based simulator of next-generation sequencing data.
McElroy KE, Luciani F, Thomas T. McElroy KE, et al. BMC Genomics. 2012 Feb 15;13:74. doi: 10.1186/1471-2164-13-74. BMC Genomics. 2012. PMID: 22336055 Free PMC article. - Substantial biases in ultra-short read data sets from high-throughput DNA sequencing.
Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Dohm JC, et al. Nucleic Acids Res. 2008 Sep;36(16):e105. doi: 10.1093/nar/gkn425. Epub 2008 Jul 26. Nucleic Acids Res. 2008. PMID: 18660515 Free PMC article. - MiSeq: A Next Generation Sequencing Platform for Genomic Analysis.
Ravi RK, Walton K, Khosroheidari M. Ravi RK, et al. Methods Mol Biol. 2018;1706:223-232. doi: 10.1007/978-1-4939-7471-9_12. Methods Mol Biol. 2018. PMID: 29423801 Review. - Bioinformatics tools and databases for analysis of next-generation sequence data.
Lee HC, Lai K, Lorenc MT, Imelfort M, Duran C, Edwards D. Lee HC, et al. Brief Funct Genomics. 2012 Jan;11(1):12-24. doi: 10.1093/bfgp/elr037. Epub 2011 Dec 19. Brief Funct Genomics. 2012. PMID: 22184335 Review.
Cited by
- Bioinformatic Amplicon Read Processing Strategies Strongly Affect Eukaryotic Diversity and the Taxonomic Composition of Communities.
Majaneva M, Hyytiäinen K, Varvio SL, Nagai S, Blomster J. Majaneva M, et al. PLoS One. 2015 Jun 5;10(6):e0130035. doi: 10.1371/journal.pone.0130035. eCollection 2015. PLoS One. 2015. PMID: 26047335 Free PMC article. - Tumor DNA as a Cancer Biomarker through the Lens of Colorectal Neoplasia.
Cohen JD, Diergaarde B, Papadopoulos N, Kinzler KW, Schoen RE. Cohen JD, et al. Cancer Epidemiol Biomarkers Prev. 2020 Dec;29(12):2441-2453. doi: 10.1158/1055-9965.EPI-20-0549. Epub 2020 Oct 8. Cancer Epidemiol Biomarkers Prev. 2020. PMID: 33033144 Free PMC article. - Accurate detection and genotyping of SNPs utilizing population sequencing data.
Bansal V, Harismendy O, Tewhey R, Murray SS, Schork NJ, Topol EJ, Frazer KA. Bansal V, et al. Genome Res. 2010 Apr;20(4):537-45. doi: 10.1101/gr.100040.109. Epub 2010 Feb 11. Genome Res. 2010. PMID: 20150320 Free PMC article. - Host-mediated microbiome engineering (HMME) of drought tolerance in the wheat rhizosphere.
Jochum MD, McWilliams KL, Pierson EA, Jo YK. Jochum MD, et al. PLoS One. 2019 Dec 4;14(12):e0225933. doi: 10.1371/journal.pone.0225933. eCollection 2019. PLoS One. 2019. PMID: 31800619 Free PMC article. - Single Nucleotide Polymorphism (SNP) Detection and Genotype Calling from Massively Parallel Sequencing (MPS) Data.
Li Y, Chen W, Liu EY, Zhou YH. Li Y, et al. Stat Biosci. 2013 May;5(1):3-25. doi: 10.1007/s12561-012-9067-4. Stat Biosci. 2013. PMID: 24489615 Free PMC article.
References
- Pennisi E. Science. 2007;318:1842–1843. - PubMed
- Chi KR. Nat. Methods. 2008;5:11–14. - PubMed
- Hillier LW, et al. Nat. Methods. 2008;5:183–188. - PubMed
Publication types
MeSH terms
Grants and funding
- P01 CA013106/CA/NCI NIH HHS/United States
- P01 CA013106-37/CA/NCI NIH HHS/United States
- P01 CA013106-38/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources