Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development - PubMed (original) (raw)
Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development
Y Jeffrey Chiang et al. PLoS One. 2008.
Abstract
Tankyrases are proteins with poly(ADP-ribose) polymerase activity. Human tankyrases post-translationally modify multiple proteins involved in processes including maintenance of telomere length, sister telomere association, and trafficking of glut4-containing vesicles. To date, however, little is known about in vivo functions for tankyrases. We recently reported that body size was significantly reduced in mice deficient for tankyrase 2, but that these mice otherwise appeared developmentally normal. In the present study, we report generation of tankyrase 1-deficient and tankyrase 1 and 2 double-deficient mice, and use of these mutant strains to systematically assess candidate functions of tankyrase 1 and tankyrase 2 in vivo. No defects were observed in development, telomere length maintenance, or cell cycle regulation in tankyrase 1 or tankyrase 2 knockout mice. In contrast to viability and normal development of mice singly deficient in either tankyrase, deficiency in both tankyrase 1 and tankyrase 2 results in embryonic lethality by day 10, indicating that there is substantial redundancy between tankyrase 1 and tankyrase 2, but that tankyrase function is essential for embryonic development.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
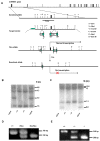
Figure 1. Generation of TANK1-deficient mice by gene targeting.
(A) Gene targeting strategy and restriction map of TANK1 gene. Filled boxes indicate exons; labeled boxes indicate neomycin (neo) resistance or herpesvirus thymidine kinase (tk) genes; and arrows indicate loxP sites. (B, C) Southern blot analysis of ES cell DNA. The 4.2 kb and 1.5 kb bands represents the germ line alleles and 6.0 kb and 2.5 kb bands represent the targeted alleles when BamHI/XbaI enzymes were used (B). The 12 kb and 1.3 kb bands represent the germ line alleles, and 13.8 kb and 0.8 kb bands represent the targeted alleles when EcoRV was used(c). (D) PCR analysis for the conditional TANK1 knockout mouse genotype. The 340- and 240-bp PCR products represent the wild-type and floxed alleles, respectively. (E) PCR analysis for the constitutive TANK1 knockout mouse genotype. The 340- and 200-bp PCR products represented the wild-type and knockout alleles, respectively.
Figure 2. Expression of TANK1 and TANK1a in WT and TANK1−/− mouse tissues.
All RNA samples were serially diluted as indicated for PCR amplification and analysis. Actin cDNA was used as an RT-PCR loading control in all experiments. (A) RT-PCR analysis for TANK1 mRNA expression in various tissues of WT and TANK1−/− (KO) mice as indicated. 5′-TANK1 RT-PCR products represent upstream cDNA of TANK1. (B) RT-PCR analysis for TANK1 mRNA expression in WT and TANK1−/− (KO) testis as indicated. 5′-TANK1 and 3′-TANK1 RT-PCR products represent upstream and downstream cDNAs of TANK1; and TANK1a RT-PCR products are specific for TANK1a. (C) RT-PCR analysis for TANK1a mRNA expression in various tissues of WT mice as indicated. TANK1a RT-PCR products represent cDNA of TANK1a. (D, E) Western blot analysis was used to determine tankyrase 1 and 1a expression in thymus, testis and spleen of WT and TANK1−/− (KO) mice with 762 (anti-SAM, D) and 376 (anti-HPS, E) antibodies. TANK1 indicates tankyrase 1 protein, TANK1x indicates a possible degraded tankyrase 1 protein, and TANK1a indicates tankyrase 1a protein.
Figure 3. Genomic structure, mRNA and protein product for TANK1 and TANK1a.
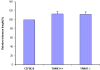
Figure 4. Telomere length was not altered in TANK1 knockout mice.
Spleen cells were isolated from C57BL/6, TANK1+/+ (n = 7) and TANK1−/− (n = 7) mice, and relative telomere length was determined by Flow-FISH. The FITC fluorescent signal of the cell-binding telomeric probe was converted to arbitrary units of molecule equivalents of soluble fluorescence (MSEF). The average of fluorescent intensities from each mouse was normalized to that of a C57BL/6 mouse (defined as 100). The relative telomere length of each strain of mice is plotted.
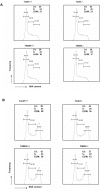
Figure 5. Cell cycle progression is equivalent in wild-type, TANK1−/−, and TANK2−/− mice.
(A) Spleen cells from TANK1−/−, TANK2−/− mice and their littermates were stimulated with IL-2, ConA and LPS for 48 hours, then fixed and stained with PI. The results shown are representative of 3 independent analyses. The percentages of G1, S and G2/M phase are indicated. (B) Spleen cells from TANK1−/−, TANK2−/− mice and their littermates were stimulated with IL-2, ConA and LPS for 48 hours, gamma irradiated at the 24 hour time point, then fixed and stained with PI. The results shown are representative of 3 independent analyses. The percentages of G1, S and G2/M phase are indicated.
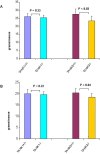
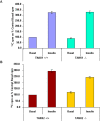
Figure 6. The body weights of tankyrase 1 knockout mice are normal while the body weights of tankyrase 2 knockout mice are reduced.
(A) Body weights in male TANK1+/+ (n = 8), TANK1−/− (n = 8), TANK2+/+ (n = 5) and TANK2−/− (n = 5) mice. (B) Body weights in female TANK1+/+ (n = 5), TANK1−/− (n = 5), TANK2+/+ (n = 4) and TANK2−/− (n = 4) mice.
Figure 7. The sensitivity of insulin-stimulated glucose uptake in tankyrase 1 knockout WAT is normal while the sensitivity of insulin-stimulated glucose uptake in tankyrase 2 knockout WAT is decreased.
(A) Assay for insulin-responsive glucose uptake in TANK1+/+and TANK1−/− WAT cells. The results shown represent three independent experiment, each using two male TANK1+/+ and 2 male TANK1−/− mice. (B) Assay for insulin-responsive glucose uptake in TANK2+/+and TANK2−/− WAT cells. The results shown represent five independent experiments, each using two male TANK2+/+ and 2 male TANK2−/− mice.
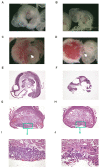
Figure 8. Abnormal phenotypes in TANK1−/−.TANK2−/− mouse embryos and placentas.
(A, C) wild-type and (B, D) TANK1−/−.TANK2−/− E10.5 embryos and placentas. Arrow points to umbilical cord. Histological sections of wild-type (E) and TANK1−/−.TANK2−/− (F) E10.5 embryos. Transverse sections of E10.5 wild-type (G) and TANK1−/−.TANK2−/− (H) placentas. The box area in (G) and (H) are shown at a higher magnification in (I) and (J), respectively.
Similar articles
- Generation and characterization of telomere length maintenance in tankyrase 2-deficient mice.
Chiang YJ, Nguyen ML, Gurunathan S, Kaminker P, Tessarollo L, Campisi J, Hodes RJ. Chiang YJ, et al. Mol Cell Biol. 2006 Mar;26(6):2037-43. doi: 10.1128/MCB.26.6.2037-2043.2006. Mol Cell Biol. 2006. PMID: 16507984 Free PMC article. - Tankyrase 2 poly(ADP-ribose) polymerase domain-deleted mice exhibit growth defects but have normal telomere length and capping.
Hsiao SJ, Poitras MF, Cook BD, Liu Y, Smith S. Hsiao SJ, et al. Mol Cell Biol. 2006 Mar;26(6):2044-54. doi: 10.1128/MCB.26.6.2044-2054.2006. Mol Cell Biol. 2006. PMID: 16507985 Free PMC article. - Insulin-stimulated exocytosis of GLUT4 is enhanced by IRAP and its partner tankyrase.
Yeh TY, Sbodio JI, Tsun ZY, Luo B, Chi NW. Yeh TY, et al. Biochem J. 2007 Mar 1;402(2):279-90. doi: 10.1042/BJ20060793. Biochem J. 2007. PMID: 17059388 Free PMC article. - Pleiotropic roles of tankyrase/PARP proteins in the establishment and maintenance of human naïve pluripotency.
Zimmerlin L, Zambidis ET. Zimmerlin L, et al. Exp Cell Res. 2020 May 1;390(1):111935. doi: 10.1016/j.yexcr.2020.111935. Epub 2020 Mar 7. Exp Cell Res. 2020. PMID: 32151493 Free PMC article. Review. - Tankyrase: a promising therapeutic target with pleiotropic action.
Sagathia V, Patel C, Beladiya J, Patel S, Sheth D, Shah G. Sagathia V, et al. Naunyn Schmiedebergs Arch Pharmacol. 2023 Dec;396(12):3363-3374. doi: 10.1007/s00210-023-02576-5. Epub 2023 Jun 20. Naunyn Schmiedebergs Arch Pharmacol. 2023. PMID: 37338576 Review.
Cited by
- Hypermetabolism, hyperphagia, and reduced adiposity in tankyrase-deficient mice.
Yeh TY, Beiswenger KK, Li P, Bolin KE, Lee RM, Tsao TS, Murphy AN, Hevener AL, Chi NW. Yeh TY, et al. Diabetes. 2009 Nov;58(11):2476-85. doi: 10.2337/db08-1781. Epub 2009 Aug 3. Diabetes. 2009. PMID: 19651815 Free PMC article. - Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Qiu W, Lam R, Voytyuk O, Romanov V, Gordon R, Gebremeskel S, Vodsedalek J, Thompson C, Beletskaya I, Battaile KP, Pai EF, Rottapel R, Chirgadze NY. Qiu W, et al. Acta Crystallogr D Biol Crystallogr. 2014 Oct;70(Pt 10):2740-53. doi: 10.1107/S1399004714017660. Epub 2014 Sep 27. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 25286857 Free PMC article. - E7449: A dual inhibitor of PARP1/2 and tankyrase1/2 inhibits growth of DNA repair deficient tumors and antagonizes Wnt signaling.
McGonigle S, Chen Z, Wu J, Chang P, Kolber-Simonds D, Ackermann K, Twine NC, Shie JL, Miu JT, Huang KC, Moniz GA, Nomoto K. McGonigle S, et al. Oncotarget. 2015 Dec 1;6(38):41307-23. doi: 10.18632/oncotarget.5846. Oncotarget. 2015. PMID: 26513298 Free PMC article. - Anti-TANKyrase weapons promote myelination.
Casaccia P. Casaccia P. Nat Neurosci. 2011 Jul 26;14(8):945-7. doi: 10.1038/nn.2885. Nat Neurosci. 2011. PMID: 21792189 Free PMC article. - Tankyrases maintain homeostasis of intestinal epithelium by preventing cell death.
Ye P, Chiang YJ, Qi Z, Li Y, Wang S, Liu Y, Li X, Chen YG. Ye P, et al. PLoS Genet. 2018 Sep 27;14(9):e1007697. doi: 10.1371/journal.pgen.1007697. eCollection 2018 Sep. PLoS Genet. 2018. PMID: 30260955 Free PMC article.
References
- Kuimov AN, Kuprash DV, Petrov VN, Vdovichenko KK, Scanlan MJ, et al. Cloning and characterization of TNKL, a member of tankyrase gene family. Genes Immun. 2001;2:52–55. - PubMed
- Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. - PubMed
- Sbodio JI, Chi NW. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J Biol Chem. 2002;277:31887–31892. - PubMed
- Seimiya H, Smith S. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182). J Biol Chem. 2002;277:14116–14126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA95099/CA/NCI NIH HHS/United States
- T32 GM007238/GM/NIGMS NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 CA095099/CA/NCI NIH HHS/United States
- GM07238/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases