Data mining of metal ion environments present in protein structures - PubMed (original) (raw)
Data mining of metal ion environments present in protein structures
Heping Zheng et al. J Inorg Biochem. 2008 Sep.
Abstract
Analysis of metal-protein interaction distances, coordination numbers, B-factors (displacement parameters), and occupancies of metal-binding sites in protein structures determined by X-ray crystallography and deposited in the PDB shows many unusual values and unexpected correlations. By measuring the frequency of each amino acid in metal ion-binding sites, the positive or negative preferences of each residue for each type of cation were identified. Our approach may be used for fast identification of metal-binding structural motifs that cannot be identified on the basis of sequence similarity alone. The analysis compares data derived separately from high and medium-resolution structures from the PDB with those from very high-resolution small-molecule structures in the Cambridge Structural Database (CSD). For high-resolution protein structures, the distribution of metal-protein or metal-water interaction distances agrees quite well with data from CSD, but the distribution is unrealistically wide for medium (2.0-2.5A) resolution data. Our analysis of cation B-factors versus average B-factors of atoms in the cation environment reveals substantial numbers of structures contain either an incorrect metal ion assignment or an unusual coordination pattern. Correlation between data resolution and completeness of the metal coordination spheres is also found.
Figures
Figure 1
Calcium-oxygen and magnesium-oxygen distance distributions for complete and incomplete coordination spheres. (A) Calcium-oxygen distance distribution for incomplete coordination spheres (CN<5) and (C) calcium-oxygen distance distribution for complete coordination spheres (CN≥5). Magnesium-oxygen distance distributions for incomplete and complete coordination spheres are shown in B and D respectively. The vertical axes give the number of interactions and the horizontal axes the distance between metal and oxygen.
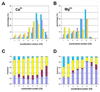
Figure 2
Metal-oxygen coordination sphere for various resolutions and coordination sphere components (calcium A,C and magnesium B,D). A,B: The horizontal axes give the coordination number and the vertical axes the percentage of structures for each data set. The cyan bars (left) correspond to structures with a resolution of 1.5Å or better, the violet bars (middle) correspond to structures with resolution between 2.0Å and 2.5Å, and the yellow bars (right) correspond to structures with a resolution worse than 2.5Å. C,D: the relative fractions of coordination sphere components. Cyan fractions (top) correspond to interaction with water, magenta (3rd from top, for 2–8) to bidentate coordination from a carboxyl group from Asp/Glu, yellow (2nd from top) to non-bidentate interaction with amino acid oxygen, and violet (bottom) to interaction with oxygen from a non-proteinaceous ligand (see the online version of this article for the colors).
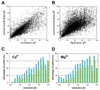
Figure 3
Scatter plots of mean _B_-factor of coordinating oxygens _versus B_-factor for (A) calcium and (B) magnesium ions. The histograms show the percentage of _B_-factor difference outliers for (C) calcium and (D) magnesium as a function of resolution. The cyan bars (left) show the percentage of points where the difference between the metal _B_-factor and the mean _B_-factor of its coordinating atoms is bigger than 5 Å2. The yellow bars (right) show the percentage of points where the difference is bigger than 10 Å2 (see the online version of this article for the colors).
Figure 4
Distributions of calcium-to-protein-oxygen and calcium-to-water distances for different resolution ranges. The vertical axes give the number of interactions in each distance bin and the horizontal axes give the distance between calcium and oxygen. Distributions are made for both oxygen from protein (A, C, E) and oxygen from water (B, D, F). Data from the CSD (A, B), PDB high resolution data (C, D), and PDB moderate resolution data (E, F) are plotted individually.
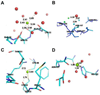
Figure 5
Unusual metal atom model parameters. (A) An atom identified as magnesium with unusually long Mg-O distances (PDB code: 1JUB; Mg A850). (B) (C) Two atoms identified as magnesium in a structure with multiple geometry problems (PDB code: 1Q9Q) (see the online version of this article for the colors).
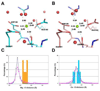
Figure 6
Re-interpretation of a magnesium binding site as calcium (PDB code: 2AS8; Mg 1001). (A) The binding site of an atom identified as magnesium with unusually long Mg-O distances. (B) Re-refinement of the same structure, after identifying the metal atom as calcium. The histograms below show the distance distributions of the metal binding site before (C) and after (D) re-interpretation. The vertical axes give the percentage of structures and the horizontal axes the metal-oxygen distance in Å. The blue lines (diamond) represent the Mg-O (C) or Ca-O (D) distance distributions for CSD data. The magenta lines (square) represent the distance distributions for high resolution PDB data. The orange bars are the magnesium-oxygen distances in 2AS8 structure (C), while the cyan bars are the calcium-oxygen distances after re-interpretation of the structure (D) (see the online version of this article for the colors).
Similar articles
- Metals in proteins: correlation between the metal-ion type, coordination number and the amino-acid residues involved in the coordination.
Dokmanić I, Sikić M, Tomić S. Dokmanić I, et al. Acta Crystallogr D Biol Crystallogr. 2008 Mar;64(Pt 3):257-63. doi: 10.1107/S090744490706595X. Epub 2008 Feb 20. Acta Crystallogr D Biol Crystallogr. 2008. PMID: 18323620 - A database overview of metal-coordination distances in metalloproteins.
Bazayeva M, Andreini C, Rosato A. Bazayeva M, et al. Acta Crystallogr D Struct Biol. 2024 May 1;80(Pt 5):362-376. doi: 10.1107/S2059798324003152. Epub 2024 Apr 29. Acta Crystallogr D Struct Biol. 2024. PMID: 38682667 Free PMC article. - Small revisions to predicted distances around metal sites in proteins.
Harding MM. Harding MM. Acta Crystallogr D Biol Crystallogr. 2006 Jun;62(Pt 6):678-82. doi: 10.1107/S0907444906014594. Epub 2006 May 12. Acta Crystallogr D Biol Crystallogr. 2006. PMID: 16699196 - Computational approaches for de novo design and redesign of metal-binding sites on proteins.
Akcapinar GB, Sezerman OU. Akcapinar GB, et al. Biosci Rep. 2017 Mar 27;37(2):BSR20160179. doi: 10.1042/BSR20160179. Print 2017 Apr 28. Biosci Rep. 2017. PMID: 28167677 Free PMC article. Review. - Structural characteristics of protein binding sites for calcium and lanthanide ions.
Pidcock E, Moore GR. Pidcock E, et al. J Biol Inorg Chem. 2001 Jun;6(5-6):479-89. doi: 10.1007/s007750100214. J Biol Inorg Chem. 2001. PMID: 11472012 Review.
Cited by
- Structural and biochemical characterization of a novel aminopeptidase from human intestine.
Tykvart J, Bařinka C, Svoboda M, Navrátil V, Souček R, Hubálek M, Hradilek M, Šácha P, Lubkowski J, Konvalinka J. Tykvart J, et al. J Biol Chem. 2015 May 1;290(18):11321-36. doi: 10.1074/jbc.M114.628149. Epub 2015 Mar 9. J Biol Chem. 2015. PMID: 25752612 Free PMC article. - Crystal structures of putative phosphoglycerate kinases from B. anthracis and C. jejuni.
Zheng H, Filippova EV, Tkaczuk KL, Dworzynski P, Chruszcz M, Porebski PJ, Wawrzak Z, Onopriyenko O, Kudritska M, Grimshaw S, Savchenko A, Anderson WF, Minor W. Zheng H, et al. J Struct Funct Genomics. 2012 Mar;13(1):15-26. doi: 10.1007/s10969-012-9131-9. Epub 2012 Mar 10. J Struct Funct Genomics. 2012. PMID: 22403005 Free PMC article. - Structure of the host-recognition device of Staphylococcus aureus phage ϕ11.
Koç C, Xia G, Kühner P, Spinelli S, Roussel A, Cambillau C, Stehle T. Koç C, et al. Sci Rep. 2016 Jun 10;6:27581. doi: 10.1038/srep27581. Sci Rep. 2016. PMID: 27282779 Free PMC article. - Amino acid influence on copper binding to peptides: cysteine versus arginine.
Wu Z, Fernandez-Lima FA, Russell DH. Wu Z, et al. J Am Soc Mass Spectrom. 2010 Apr;21(4):522-33. doi: 10.1016/j.jasms.2009.12.020. Epub 2010 Jan 11. J Am Soc Mass Spectrom. 2010. PMID: 20138783 - fingeRNAt-A novel tool for high-throughput analysis of nucleic acid-ligand interactions.
Szulc NA, Mackiewicz Z, Bujnicki JM, Stefaniak F. Szulc NA, et al. PLoS Comput Biol. 2022 Jun 2;18(6):e1009783. doi: 10.1371/journal.pcbi.1009783. eCollection 2022 Jun. PLoS Comput Biol. 2022. PMID: 35653385 Free PMC article.
References
- Engh R, Huber R. Acta Cryst. A. 1991;47:392–400.
- Jaskolski M, Gilski M, Dauter Z, Wlodawer A. Acta Cryst. D. 2007;63:611–620. - PubMed
- Harding M. Acta Cryst. D. 2001;57:401–411. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM53163/GM/NIGMS NIH HHS/United States
- R01 GM053163/GM/NIGMS NIH HHS/United States
- GM74942/GM/NIGMS NIH HHS/United States
- U54 GM074942-010006/GM/NIGMS NIH HHS/United States
- R01 GM053163-13/GM/NIGMS NIH HHS/United States
- U54 GM074942/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources