Dap160/intersectin binds and activates aPKC to regulate cell polarity and cell cycle progression - PubMed (original) (raw)
Dap160/intersectin binds and activates aPKC to regulate cell polarity and cell cycle progression
Chiswili Chabu et al. Development. 2008 Aug.
Abstract
The atypical protein kinase C (aPKC) is required for cell polarization of many cell types, and is upregulated in several human tumors. Despite its importance in cell polarity and growth control, relatively little is known about how aPKC activity is regulated. Here, we use a biochemical approach to identify Dynamin-associated protein 160 (Dap160; related to mammalian intersectin) as an aPKC-interacting protein in Drosophila. We show that Dap160 directly interacts with aPKC, stimulates aPKC activity in vitro and colocalizes with aPKC at the apical cortex of embryonic neuroblasts. In dap160 mutants, aPKC is delocalized from the neuroblast apical cortex and has reduced activity, based on its inability to displace known target proteins from the basal cortex. Both dap160 and aPKC mutants have fewer proliferating neuroblasts and a prolonged neuroblast cell cycle. We conclude that Dap160 positively regulates aPKC activity and localization to promote neuroblast cell polarity and cell cycle progression.
Figures
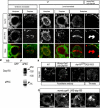
Fig. 1. Dap160 interacts with aPKC
(A) Drosophila Dap160 and vertebrate intersectin protein domains. (B) Dap160 antibody detects two bands in wild-type lysate (top arrow, 160 kDa; bottom arrow, 120 kDa) that may correspond to the two predicted isoforms shown in A; and these bands are absent in lysate from dap160 mutants, illustrating the specificity of the antibody. An independently generated antibody gives the same result (see Fig. 2N). (C) Immunoprecipitation from larval lysate using an aPKC antibody and a control antibody (Bgal) and blotted with a Dap160 antibody shows aPKC co-immunoprecipitates Dap160 protein (arrowhead). (D,E) In vitro protein interaction experiments. (D) In vitro generated Dap160 protein coupled to glutathione S-transferase (GST) beads can bind in vitro produced aPKC protein (arrowhead). Beads alone do not bind aPKC; input lane shown at left. (E) In vitro generated Dap160 protein coupled to glutathione S-transferase (GST) beads can bind in vitro produced Par-6 protein (arrowhead). Beads alone do not bind Par-6; input lane shown at left. (F,G) Dap160 directly stimulates aPKC activity and this effect can be partially blocked by Par6. Top: presence (+) or absence (−) of each protein; middle: phosphorylation of the aPKC substrate peptide. Histogram shows quantification of the phosphorylation (bars) over a schematic depiction of protein levels (see Materials and methods for protein concentrations). Note that aPKC alone can have high activity immediately after its synthesis (F) or much lower activity after storage (G), but can still be stimulated by Dap160.
Fig. 2. Dap160 co-localizes with aPKC in neuroblasts
(A-L) Neuroblasts co-stained for Dap160 (A-F) and aPKC (G-L); merged images below. Genotypes, developmental stages and cell cycle stages labeled at top. (B,H) Dap160 colocalizes with aPKC at the apical cortex of mitotic embryonic neuroblasts. (E,K) Endogenous Dap160 is undetectable at the site of aPKC apical cortical localization in larval brain metaphase neuroblasts. (F,L) Overexpression of Dap160 reveals colocalization with aPKC at the apical cortex in larval brain metaphase neuroblasts (90%, _n_=12). Scale bar: 5 μm. (M) aPKC can immunoprecipitate Dap160 from embryonic lysates. Right lane: aPKC antibody can immunoprecipitate Dap160 (top arrow) and aPKC (bottom arrow). Left lane: the GFP control antibody does not immunoprecipitate either Dap160 or aPKC. (N) Two independently generated Dap160 antibodies detect apical crescents of Dap160 in wild-type neuroblasts or Dap160 overexpression neuroblasts (arrowheads) but not in dap160 mutant neuroblasts (brackets). Roos/Marie antibody from Roos and Kelly (Roos and Kelly, 1998). Scale bar: 5 μm. (O) Dap160 overexpression results in the formation of Dap160/Sec15 double-positive puncta in neuroblasts (arrowheads; left pair of panels) and Dap160/aPKC double-positive puncta (arrowheads; right pair of panels). Scale bar: 5 μm.
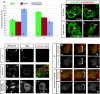
Fig. 3. Dap160 regulates embryonic neuroblast cortical polarity
(A-E) Wild type metaphase embryonic stage 15 neuroblasts stained for the indicated proteins (top labels) have apical crescents of aPKC, Bazooka, Par-6 (100%, _n_=23; 100%, _n_=34; and 96%, _n_=23, respectively) and basal crescents of Miranda (93%, _n_=43) and Numb (100%, _n_=21) (white arrowheads). (F-J) dap160 metaphase embryonic stage 15 neuroblasts. (F) aPKC is mostly ectopic cortical aPKC (71%, _n_=41, red arrow) or cytoplasmic (not shown). (G) Par-6 is ectopic cortical (50%, _n_=12, red arrow) or normal (not shown). (H) Baz is cytoplasmic (21.8%, _n_=96; H), ectopic cortical or normal (not shown). (I) Miranda is ectopic cortical (red arrow, 36%, _n_=55) or normal (not shown). (J) Numb is ectopic cortical (red arrow, 38%, _n_=18) or normal (not shown). Proteins are delocalized and may also be at lower levels than in wild-type neuroblasts. (K-M) dynamin mutant mitotic larval neuroblasts (shibirets/shibirets at restrictive temperature; see Materials and methods) have aPKC apical crescents (K) and basal crescents of Miranda (L) and Numb (M). (N) Quantification of the neuroblast cortical polarity phenotypes. Number of neuroblasts scored is shown as a number at the bottom of each bar. Scale bar: 5 μm.
Fig. 4. Dap160 positively regulates neuroblast pool size
(A) Neuroblast numbers scored wild-type (green bars), dap160 mutants (red bars) and Dap160 misexpression larvae at second instar (L2) or wandering third instar (L3). Numbers inside bars indicate numbers of brains analyzed. See Materials and methods for genotypes and growth temperatures. (B-E) Wild-type (B,C), dap160 (D) and aPKC (E) mutant clones (_n_≥50 and _n_=35 respectively) always contain a single Deadpan-positive neuroblast (arrowhead). (F-L) Neuroblast cortical polarity in larval neuroblasts. (F,G) Wild-type neuroblasts have aPKC apical crescents (arrowhead) and Miranda (Mira) basal crescents. (H,I) dap160 mutants have weak ectopic cortical aPKC (arrows) and normal Mira basal crescents (15%, _n_=13). (J,K) Dap160 overexpression in neuroblasts leads to weak ectopic cortical aPKC (arrow), increased cytoplasmic and reduced cortical Mira (19%, _n_=21 as shown; remainder normal basal crescents); Dap160 is detected in cortical patches and cytoplasmic puncta (L). (M,N) Live imaging of wild-type neuroblasts with GFP::Mira and Cherry::Jupiter. (M) Wild-type neuroblasts show basal GFP::Mira at metaphase (brackets) and partition GFP::Mira to the GMC at telophase (arrowhead; 100%, _n_=37). (N) Dap160 misexpressing neuroblasts show cytoplasmic GFP::Mira at metaphase (brackets) and occasionally do not segregate GFP::Mira to the GMC at telophase (arrow; 8%, _n_=40). Scale bars: 5 μm.
Fig. 5. Dap160 is required for neuroblast cell cycle progression
Wild-type, dap160 mutant and aPKC mutant neuroblasts imaged with Jupiter::GFP from nuclear envelope breakdown (NEBD) to anaphase onset (AO). (A) Wild-type neuroblasts have a NEBD-AO interval of 7.76±2.04 minutes; _n_=15. (B) dap160 mutant neuroblasts have a NEBD-AO interval of 13.37±4.4 minutes; _n_=10. (C) aPKC mutant neuroblasts have a NEBD-AO interval of 17.84±4.52 minutes; _n_=11. Scale bar: 5 μm.
Similar articles
- Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC.
Atwood SX, Chabu C, Penkert RR, Doe CQ, Prehoda KE. Atwood SX, et al. J Cell Sci. 2007 Sep 15;120(Pt 18):3200-6. doi: 10.1242/jcs.014902. Epub 2007 Aug 28. J Cell Sci. 2007. PMID: 17726059 Free PMC article. - Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation.
Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, Doe CQ. Lee CY, et al. Genes Dev. 2006 Dec 15;20(24):3464-74. doi: 10.1101/gad.1489406. Genes Dev. 2006. PMID: 17182871 Free PMC article. - Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia.
Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ. Rolls MM, et al. J Cell Biol. 2003 Dec 8;163(5):1089-98. doi: 10.1083/jcb.200306079. Epub 2003 Dec 1. J Cell Biol. 2003. PMID: 14657233 Free PMC article. - Polarization of Drosophila neuroblasts during asymmetric division.
Prehoda KE. Prehoda KE. Cold Spring Harb Perspect Biol. 2009 Aug;1(2):a001388. doi: 10.1101/cshperspect.a001388. Cold Spring Harb Perspect Biol. 2009. PMID: 20066083 Free PMC article. Review. - Lgl/aPKC and Crb regulate the Salvador/Warts/Hippo pathway.
Parsons LM, Grzeschik NA, Allott ML, Richardson HE. Parsons LM, et al. Fly (Austin). 2010 Oct-Dec;4(4):288-93. doi: 10.4161/fly.4.4.13116. Epub 2010 Oct 21. Fly (Austin). 2010. PMID: 20798605 Free PMC article. Review.
Cited by
- Mask, the Drosophila ankyrin repeat and KH domain-containing protein, affects microtubule stability.
Martinez D, Zhu M, Guidry JJ, Majeste N, Mao H, Yanofsky ST, Tian X, Wu C. Martinez D, et al. J Cell Sci. 2021 Oct 15;134(20):jcs258512. doi: 10.1242/jcs.258512. Epub 2021 Oct 22. J Cell Sci. 2021. PMID: 34553767 Free PMC article. - EGFR/ARF6 regulation of Hh signalling stimulates oncogenic Ras tumour overgrowth.
Chabu C, Li DM, Xu T. Chabu C, et al. Nat Commun. 2017 Mar 10;8:14688. doi: 10.1038/ncomms14688. Nat Commun. 2017. PMID: 28281543 Free PMC article. - Gene identification and RNAi-silencing of p62/SQSTM1 in the vector Rhodnius prolixus reveals a high degree of sequence conservation but no apparent deficiency-related phenotypes in vitellogenic females.
Pereira J, Santos-Araujo S, Bomfim L, Gondim KC, Majerowicz D, Pane A, Ramos I. Pereira J, et al. PLoS One. 2023 Jul 24;18(7):e0287488. doi: 10.1371/journal.pone.0287488. eCollection 2023. PLoS One. 2023. PMID: 37486954 Free PMC article. - Dynamin participates in the maintenance of anterior polarity in the Caenorhabditis elegans embryo.
Nakayama Y, Shivas JM, Poole DS, Squirrell JM, Kulkoski JM, Schleede JB, Skop AR. Nakayama Y, et al. Dev Cell. 2009 Jun;16(6):889-900. doi: 10.1016/j.devcel.2009.04.009. Dev Cell. 2009. PMID: 19531359 Free PMC article. - aPKC: the Kinase that Phosphorylates Cell Polarity.
Hong Y. Hong Y. F1000Res. 2018 Jun 25;7:F1000 Faculty Rev-903. doi: 10.12688/f1000research.14427.1. eCollection 2018. F1000Res. 2018. PMID: 29983916 Free PMC article. Review.
References
- Adams A, Thorn JM, Yamabhai M, Kay BK, O'Bryan JP. Intersectin, an adaptor protein involved in clathrin-mediated endocytosis, activates mitogenic signaling pathways. J. Biol. Chem. 2000;275:27414–27420. - PubMed
- Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, Pawson T, Muthuswamy SK. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat. Cell Biol. 2006;8:1235–1245. - PubMed
- Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 2007;9:1066–1073. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous