Drosophila RNAi screen identifies host genes important for influenza virus replication - PubMed (original) (raw)
Drosophila RNAi screen identifies host genes important for influenza virus replication
Linhui Hao et al. Nature. 2008.
Abstract
All viruses rely on host cell proteins and their associated mechanisms to complete the viral life cycle. Identifying the host molecules that participate in each step of virus replication could provide valuable new targets for antiviral therapy, but this goal may take several decades to achieve with conventional forward genetic screening methods and mammalian cell cultures. Here we describe a novel genome-wide RNA interference (RNAi) screen in Drosophila that can be used to identify host genes important for influenza virus replication. After modifying influenza virus to allow infection of Drosophila cells and detection of influenza virus gene expression, we tested an RNAi library against 13,071 genes (90% of the Drosophila genome), identifying over 100 for which suppression in Drosophila cells significantly inhibited or stimulated reporter gene (Renilla luciferase) expression from an influenza-virus-derived vector. The relevance of these findings to influenza virus infection of mammalian cells is illustrated for a subset of the Drosophila genes identified; that is, for three implicated Drosophila genes, the corresponding human homologues ATP6V0D1, COX6A1 and NXF1 are shown to have key functions in the replication of H5N1 and H1N1 influenza A viruses, but not vesicular stomatitis virus or vaccinia virus, in human HEK 293 cells. Thus, we have demonstrated the feasibility of using genome-wide RNAi screens in Drosophila to identify previously unrecognized host proteins that are required for influenza virus replication. This could accelerate the development of new classes of antiviral drugs for chemoprophylaxis and treatment, which are urgently needed given the obstacles to rapid development of an effective vaccine against pandemic influenza and the probable emergence of strains resistant to available drugs.
Figures
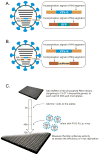
Fig. 1. Overview of genome-wide RNAi screen to identify host factors involved in influenza virus replication in Drosophila cells
Schematic diagrams showing recombinant influenza viruses, (A) FVG-G, in which genes encoding the HA and NA proteins were replaced with the VSV-G and eGFP genes, respectively, and (B) FVG-R, in which the genes encoding the HA and NA were replaced with the VSV-G and Renilla luciferase genes, respectively. (C) Schematic diagram of systematic analysis of host genes affecting influenza virus replication and gene expression in Drosophila cells. Experimental details are given in Methods.
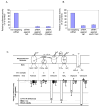
Fig. 2. Effect of selected siRNAs and inhibitors on Renilla luciferase expression in FVG-R-infected human cells
Renilla luciferase activity was measured in FVG-R-infected 293 cells treated with siRNAs against (A) ATP6V0D1, COX6A1, (B) NXF1, or (C) the indicated mitochondrial electron transport chain inhibitors. Inhibitors of complexes III, IV and V inhibited FVG-R-directed Renilla luciferase expression significantly, while complex I and II inhibitors had little or no effect. In contrast, the inhibitors had no significant effects on cell viability and Gaussia luciferase expression of a murine leukemia virus derivative (MLV-GL) that, like FVG-R, depended on VSV G-mediated entry. All experiments (A-C) were conducted three times in duplicate, with the results reported as means ± SD.
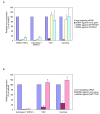
Fig. 3. Effect of siRNAs against selected genes on the replication of influenza viruses, VSV or vaccinia virus in human 293 cells
The titers of influenza viruses (WSN and Indonesia 7), VSV and vaccinia virus in 293 cells treated with siRNA against (A) ATP6V0D1, COX6A1 or (B) NXF1 are shown. Experimental details are given in Supplementary Methods. All experiments were conducted three times, with the results reported as means ± SD.
Similar articles
- Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication.
Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Mäurer AP, Müller E, Wolff T, Rudel T, Meyer TF. Karlas A, et al. Nature. 2010 Feb 11;463(7282):818-22. doi: 10.1038/nature08760. Epub 2010 Jan 17. Nature. 2010. PMID: 20081832 - Genome-wide RNAi screen for viral replication in mammalian cell culture.
Prusty BK, Karlas A, Meyer TF, Rudel T. Prusty BK, et al. Methods Mol Biol. 2011;721:383-95. doi: 10.1007/978-1-61779-037-9_24. Methods Mol Biol. 2011. PMID: 21431699 - A genome-wide RNAi screen reveals that mRNA decapping restricts bunyaviral replication by limiting the pools of Dcp2-accessible targets for cap-snatching.
Hopkins KC, McLane LM, Maqbool T, Panda D, Gordesky-Gold B, Cherry S. Hopkins KC, et al. Genes Dev. 2013 Jul 1;27(13):1511-25. doi: 10.1101/gad.215384.113. Genes Dev. 2013. PMID: 23824541 Free PMC article. - Cellular networks involved in the influenza virus life cycle.
Watanabe T, Watanabe S, Kawaoka Y. Watanabe T, et al. Cell Host Microbe. 2010 Jun 25;7(6):427-39. doi: 10.1016/j.chom.2010.05.008. Cell Host Microbe. 2010. PMID: 20542247 Free PMC article. Review. - Defense Mechanisms against Viral Infection in Drosophila: RNAi and Non-RNAi.
Swevers L, Liu J, Smagghe G. Swevers L, et al. Viruses. 2018 May 1;10(5):230. doi: 10.3390/v10050230. Viruses. 2018. PMID: 29723993 Free PMC article. Review.
Cited by
- Inhibitory and combinatorial effect of diphyllin, a v-ATPase blocker, on influenza viruses.
Chen HW, Cheng JX, Liu MT, King K, Peng JY, Zhang XQ, Wang CH, Shresta S, Schooley RT, Liu YT. Chen HW, et al. Antiviral Res. 2013 Sep;99(3):371-82. doi: 10.1016/j.antiviral.2013.06.014. Epub 2013 Jun 29. Antiviral Res. 2013. PMID: 23820269 Free PMC article. - Identification of Nifurtimox and Chrysin as Anti-Influenza Virus Agents by Clinical Transcriptome Signature Reversion.
Xin Y, Chen S, Tang K, Wu Y, Guo Y. Xin Y, et al. Int J Mol Sci. 2022 Feb 21;23(4):2372. doi: 10.3390/ijms23042372. Int J Mol Sci. 2022. PMID: 35216485 Free PMC article. - Nucleolin/Nsr1p binds to the 3' noncoding region of the tombusvirus RNA and inhibits replication.
Jiang Y, Li Z, Nagy PD. Jiang Y, et al. Virology. 2010 Jan 5;396(1):10-20. doi: 10.1016/j.virol.2009.10.007. Epub 2009 Oct 27. Virology. 2010. PMID: 19861225 Free PMC article. - KAP1 Positively Modulates Influenza A Virus Replication by Interacting with PB2 and NS1 Proteins in Human Lung Epithelial Cells.
Feng H, Yi R, Wu S, Wang G, Sun R, Lin L, Zhu S, Nie Z, He Y, Wang S, Wang P, Shu J, Wu L. Feng H, et al. Viruses. 2022 Mar 26;14(4):689. doi: 10.3390/v14040689. Viruses. 2022. PMID: 35458419 Free PMC article. - Susceptibility Genes to Plant Viruses.
Garcia-Ruiz H. Garcia-Ruiz H. Viruses. 2018 Sep 10;10(9):484. doi: 10.3390/v10090484. Viruses. 2018. PMID: 30201857 Free PMC article. Review.
References
- Kuttenkeuler D, Boutros M. Genome-wide RNAi as a route to gene function in Drosophila. Brief Funct Genomic Proteomic. 2004;3:168–76. - PubMed
- Webster RG, Govorkova EA. H5N1 influenza--continuing evolution and spread. N Engl J Med. 2006;355:2174–7. - PubMed
- Ludwig S, Planz O, Pleschka S, Wolff T. Influenza-virus-induced signaling cascades: targets for antiviral therapy? Trends Mol Med. 2003;9:46–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM035072/GM/NIGMS NIH HHS/United States
- R01 AI069274-03/AI/NIAID NIH HHS/United States
- R01 AI044386-10/AI/NIAID NIH HHS/United States
- GM35072/GM/NIGMS NIH HHS/United States
- R37 GM035072/GM/NIGMS NIH HHS/United States
- R01 AI069274/AI/NIAID NIH HHS/United States
- R01 AI044386/AI/NIAID NIH HHS/United States
- R01 AI047446/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM035072-23/GM/NIGMS NIH HHS/United States
- R01 AI047446-09/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous