Membrane fusion - PubMed (original) (raw)
Review
Membrane fusion
William Wickner et al. Nat Struct Mol Biol. 2008 Jul.
Abstract
Subcellular compartmentalization, cell growth, hormone secretion and neurotransmission require rapid, targeted, and regulated membrane fusion. Fusion entails extensive lipid rearrangements by two apposed (docked) membrane vesicles, joining their membrane proteins and lipids and mixing their luminal contents without lysis. Fusion of membranes in the secretory pathway involves Rab GTPases; their bound 'effector' proteins, which mediate downstream steps; SNARE proteins, which can 'snare' each other, in cis (bound to one membrane) or in trans (anchored to apposed membranes); and SNARE-associated proteins (SM proteins; NSF or Sec18p; SNAP or Sec17p; and others) cooperating with specific lipids to catalyze fusion. In contrast, mitochondrial and cell-cell fusion events are regulated by and use distinct catalysts.
Figures
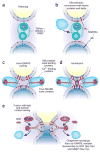
Figure 1
Membrane fusion on the exocytic and endocytic pathways, in five steps. (a) The first association of membranes, termed ‘tethering’, requires a prenyl-anchored Rab-family GTPase and tethering proteins termed ‘effectors’, which bind to the Rab in its activated, GTP-bound state. Proteins mediating tethering have been studied in the Golgi stacks, and other systems. (b) Rab-regulated enrichment of fusion proteins and lipids in a microdomain. Rabs, their multi-functional effector complexes, and lipids with defined roles in fusion (such as sterols (not shown) or phosphoinositides or diacylglycerol (red polar head groups)) assemble into a microdomain, the site of subsequent steps in the fusion pathway. In some systems, Rab effectors can include guanine nucleotide exchange factors, which activate Rabs; lipid kinases, which synthesize phosphoinositides; and SM proteins, which bind SNAREs. It remains unclear in most instances whether Rab effectors must remain bound to the Rab to be activated for these functions, or whether concentration of these several protein and lipid factors in the fusion microdomain suffices. In some systems, such as the yeast vacuole, one multisubunit complex fulfills tethering, guanine nucleotide exchange, SM protein, and lipid-binding functions. (c) Assembly of _trans_-SNARE complexes with additional regulatory proteins. These include SM proteins and can include proteins or domains that bind to Ca2+, to lipids or to SNAREs. These complexes may encircle the site of future fusion. Lipids with small head groups and negative membrane curvature, which promote hemifusion, are enriched at the cytoplasmic surface of the fusion microdomain (red head groups). (d) Hemifusion, formed by fusion of the halves of the lipid bilayer of each membrane that face the cytoplasm. Arrows indicate the direction of subsequent lipid movement to complete the fusion process. Lipids with positive curvature due to large head groups (shown here as blue head groups) may become enriched at this stage for invasion of the hemifused structure (arrows). (e) Completion of fusion, with mixing of lipid bilayers, membrane proteins and luminal compartments but retention of the barrier between cytoplasm and organellar lumen. This process converts _trans_-SNARE complexes to post-fusion _cis_-SNARE complexes; it is unclear whether _cis_-SNARE complexes can arise by any other route. SNAP (Sec17p) may displace other SNARE-bound proteins and prepare the _cis_-SNARE complex for ATP-dependent disassembly by NSF (Sec18p).
Figure 2
Mitochondrial fusion and cell fusion in mating yeast cells. (a) Mitochondria initiate contact and fusion through the interaction of Fzo1 (or Mfn in mammals), a dynamin-like GTPase located in the outer mitochondrial membrane. The fusion of the outer membrane occurs first, and then inner membrane contact and fusion is regulated by another dynamin-like GTPase, Mgm1 (or OPA1 in mammals). Ugo1 in the outer membrane provides a physical link between Fzo1 and Mgm1. (b) Yeast cells initiate cell fusion by regulated expression of two membrane proteins, Fus1p (a single-pass membrane protein) and Prm1p (a multispanning membrane protein), and a cytoplasmic protein, Fus2p. Fus1p and Fus2p localize to the cell mating tip and are required for the dissolution of the cell wall separating the plasma membranes of the mating pair. Prm1p is required for some as-yet-undefined reaction, possibly the formation of a fusion pore, that occurs when the mating cell plasma membranes come into contact.
Similar articles
- Homotypic vacuolar fusion mediated by t- and v-SNAREs.
Nichols BJ, Ungermann C, Pelham HR, Wickner WT, Haas A. Nichols BJ, et al. Nature. 1997 May 8;387(6629):199-202. doi: 10.1038/387199a0. Nature. 1997. PMID: 9144293 - A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion.
Ungermann C, Nichols BJ, Pelham HR, Wickner W. Ungermann C, et al. J Cell Biol. 1998 Jan 12;140(1):61-9. doi: 10.1083/jcb.140.1.61. J Cell Biol. 1998. PMID: 9425154 Free PMC article. - Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones.
Mima J, Hickey CM, Xu H, Jun Y, Wickner W. Mima J, et al. EMBO J. 2008 Aug 6;27(15):2031-42. doi: 10.1038/emboj.2008.139. Epub 2008 Jul 24. EMBO J. 2008. PMID: 18650938 Free PMC article. - Yeast homotypic vacuole fusion: a window on organelle trafficking mechanisms.
Wickner W, Haas A. Wickner W, et al. Annu Rev Biochem. 2000;69:247-75. doi: 10.1146/annurev.biochem.69.1.247. Annu Rev Biochem. 2000. PMID: 10966459 Review. - Functions of SNAREs in intracellular membrane fusion and lipid bilayer mixing.
Ungermann C, Langosch D. Ungermann C, et al. J Cell Sci. 2005 Sep 1;118(Pt 17):3819-28. doi: 10.1242/jcs.02561. J Cell Sci. 2005. PMID: 16129880 Review.
Cited by
- Conserved and plant-unique mechanisms regulating plant post-Golgi traffic.
Fujimoto M, Ueda T. Fujimoto M, et al. Front Plant Sci. 2012 Aug 28;3:197. doi: 10.3389/fpls.2012.00197. eCollection 2012. Front Plant Sci. 2012. PMID: 22973281 Free PMC article. - The delivery of therapeutic oligonucleotides.
Juliano RL. Juliano RL. Nucleic Acids Res. 2016 Aug 19;44(14):6518-48. doi: 10.1093/nar/gkw236. Epub 2016 Apr 15. Nucleic Acids Res. 2016. PMID: 27084936 Free PMC article. Review. - The comings and goings of MHC class I molecules herald a new dawn in cross-presentation.
Blander JM. Blander JM. Immunol Rev. 2016 Jul;272(1):65-79. doi: 10.1111/imr.12428. Immunol Rev. 2016. PMID: 27319343 Free PMC article. Review. - Physico-Chemical Mechanisms of the Functioning of Membrane-Active Proteins of Enveloped Viruses.
Batishchev OV. Batishchev OV. Biochem (Mosc) Suppl Ser A Membr Cell Biol. 2022;16(4):247-260. doi: 10.1134/S1990747822050038. Epub 2022 Dec 9. Biochem (Mosc) Suppl Ser A Membr Cell Biol. 2022. PMID: 36532264 Free PMC article. - Salmonella SipA mimics a cognate SNARE for host Syntaxin8 to promote fusion with early endosomes.
Singh PK, Kapoor A, Lomash RM, Kumar K, Kamerkar SC, Pucadyil TJ, Mukhopadhyay A. Singh PK, et al. J Cell Biol. 2018 Dec 3;217(12):4199-4214. doi: 10.1083/jcb.201802155. Epub 2018 Oct 11. J Cell Biol. 2018. PMID: 30309979 Free PMC article.
References
- Siegel DP, et al. Physiological levels of diacylglycerols in phospholipid membranes induce membrane fusion and stabilize inverted phases. Biochemistry. 1989;38:3703–3709. - PubMed
- Burgess SW, McIntosh TJ, Lentz BR. Modulation of poly(ethylene glycol)-induced fusion by membrane hydration: importance of interbilayer separation. Biochemistry. 1992;31:2653–2661. - PubMed
- Lee J, Lentz BR. Evolution of lipidic structures during model membrane fusion and the relation of this process to cell membrane fusion. Biochemistry. 1997;36:6251–6259. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources