MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation - PubMed (original) (raw)
MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation
Jenn-Yah Yu et al. Exp Cell Res. 2008.
Abstract
MicroRNAs (miRNAs) are small RNAs with diverse regulatory roles. The miR-124 miRNA is expressed in neurons in the developing and adult nervous system. Here we show that overexpression of miR-124 in differentiating mouse P19 cells promotes neurite outgrowth, while blocking miR-124 function delays neurite outgrowth and decreases acetylated alpha-tubulin. Altered neurite outgrowth also was observed in mouse primary cortical neurons when miR-124 expression was increased, or when miR-124 function was blocked. In uncommitted P19 cells, miR-124 expression led to disruption of actin filaments and stabilization of microtubules. Expression of miR-124 also decreased Cdc42 protein and affected the subcellular localization of Rac1, suggesting that miR-124 may act in part via alterations to members of the Rho GTPase family. Furthermore, constitutively active Cdc42 or Rac1 attenuated neurite outgrowth promoted by miR-124. To obtain a broader perspective, we identified mRNAs downregulated by miR-124 in P19 cells using microarrays. mRNAs for proteins involved in cytoskeletal regulation were enriched among mRNAs downregulated by miR-124. A miR-124 variant with an additional 5' base failed to promote neurite outgrowth and downregulated substantially different mRNAs. These results indicate that miR-124 contributes to the control of neurite outgrowth during neuronal differentiation, possibly by regulation of the cytoskeleton.
Figures
Figure 1
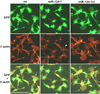
(A) Northern blot analysis detects mature miR-124 in mouse P19 cells transiently transfected with MASH1 or Ngn2 expression vectors. miR-124 is also detected in P19 cells transfected with expression vectors for three different mouse miR-124 genes (miR-124-1, miR-124-2, and miR-124-3), or a vector expressing a 3.8 Kb cDNA for miR-124-1, but not from a mutant form of miR-124-1 (miR-124-1m). Blots were stripped and reprobed for the U6 snRNA as loading control. (B) P19 cells were transfected with 2’-O-Me RNA oligos complementary to miR-124 (O-Me-124-as) or a control scrambled miR-124 sequence (O-Me-124-sc), in combination with plasmids expressing GFP and MASH1. Cells were fixed at 57 hr after transfection and stained with anti-GFP and TuJ1 antibodies. (C) Quantification of neurite initiation for 2-O-Me-124-as or 2-O-Me-124-sc transfected cells at 57 hr. GFP positive cells with processes longer than 2 cell-body diameters were considered as neurons with neurites. Average percentage of GFP positive neurons with neurites is shown, with standard error indicated. ** p<0.01, Student’s t-test. (D–F) miR-124 expression vectors or a control expression vector expressing a myc-epitope tag (mt) were cotransfected with plasmids expressing GFP and MASH1. Transfected P19 cells were fixed 46 hr after transfection, stained with anti-GFP and TuJ1 antibodies, and neurite initiation was quantitated (D). Experiments were done in three repeats. Average percentage of GFP positive neurons with neurites with standard error indicated. ** p<0.01, Student’s t-test. (E) Enhanced neurite outgrowth in cells cotransfected with miR-124-1 vector, but not with the miR-124-1m vector. (D) Transfected cells fixed at 64 hr after transfection and stained with anti-GFP and TuJ1 antibodies.
Figure 2. A synthetic miR-124 RNA duplex is sufficient to promote neurite outgrowth
(A) Schematic graph shows a luciferase sensor construct used to assess miRNA transfection and function. One copy of a sequence completely complementary to miR-124 and miR-124-UU21/22 is present after the luciferase coding region. (B) Sequences of synthetic miRNA duplexes are shown in the first column. Presence or absence of the enhanced neurite outgrowth phenotype as described in Fig. 1 is indicated. Average relative luciferase activities for each miRNA duplex cotransfected with the sensor construct in A were measured at 22 hr after transfection is shown ± standard error. ** p<0.01, Student’s t-test.
Figure 3
miR-124 expression alters the morphology and actin cytoskeleton of P19 cells. P19 cells were cotransfected with the miR-124-1 expression vector, the miR-124-1m expression vector or the mt control vector in combination with plasmids expressing GFP (green) and puromycin resistance genes. Transfected cells were selected in puromycin, fixed at 44 hr after transfection and stained with Alexa-546-conjugated phalloidin (red). Inset shows an example of fine F-actin containing processes frequent at the edges of miR-124 expressing cells (position of inset indicated by arrowhead). Arrows indicate lamellipodia in the control and miR-124-1m transfections. Scale bar = 100µm.
Figure 4
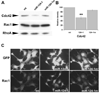
Actin, F-actin, and acetylated α-tubulin levels in miR-124 expressing P19 cells. Transfected cells were cultured and selected as described for Fig. 3. For A–C, proteins were extracted from transfected P19 cells 44 hour after transfection. (A) Western blot analysis of total actin, acetylated α-tubulin and α-tubulin. (B) Quantification of western blot analysis in A. Density of the bands on X-ray films was scanned, quantified by Image J. (C) Quantification of methanol extracted F-actin bound to Alexa-546 conjugated phalloidin. ( D) Quantification of western blot analyses for acetylated α-tubulin and α-tubulin in MASH1 transfected P19 cells in which miR-124 function was blocked with 2-O-Me-124-as oligonucleotides. α-tubulin levels are reduced relative to the 2-O-Me-124-sc control. Proteins were extracted from the transfected cells at 48 hr after transfection. Average densities are shown with standard error indicated. * p<0.05, ** p<0.01, Student’s t-test.
Figure 5. miR-124 alters Cdc42 levels and Rac1 localization
(A) Western blot analysis of Cdc42, Rac1, and RhoA. (B) Quantification of Cdc42 levels in (A). Bar graph is shown as average with standard error indicated as error bar. ** p< 0.01, Student’s t-test. (C) miR-124 increases nuclear accumulation of Rac1. Transfected P19 cells were fixed at 44 hr after transfection and processed with antibodies against GFP and Rac1.
Figure 6
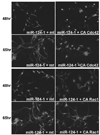
Constitutively active forms of Cdc42 or Rac1 inhibit neurite outgrowth promoted by miR-124 in MASH1 transfected P19 cells. Cdc42V12 (CA-Cdc42), Rac1V12 (CA-Rac1), or mt control expression constructs was co-transfected with plasmids expressing miR-124 (124-1), GFP, and MASH1. Transfected cells were allowed to differentiate as described for Fig. 1. P19 cells were transfected with mt or CA-Cdc42 in (A), mt or CA-Rac1 in (B). Cells were fixed at 48hr or 64 hr as indicated after transfection and processed with the TuJ1 antibody.
Figure 7
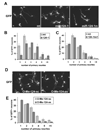
Expression of miR-124 increases number of primary neurites extended from cell bodies of cortical neurons and blocking of miR-124 decreases number of primary neurites. Cortical progenitors freshly dissociated from E14.5 cortex were transfected with GFP plasmid and different expression constructs or 2’-O-Me oligos as indicated. Cells were cultured for 43 hr in vitro before fixation, cells were processed for indirect immunofluorescence with an antibody to GFP, and neurite outgrowth was analyzed. (A) Cortical neurons transfected with mt, 124-1, and 124-1m. (B) Quantification of primary neurites extended from the transfected neurons. GFP positive neurons were categorized according to number of primary neurites (processes longer than one cell-body diameter) extended directly from their cell bodies. Histograms of 124-1 group and control (mt) group were plotted side by side. miR-124-1 shifted the distribution toward fewer neuritis per cell. Difference of the two distributions is significant according to Wilcoxon’s rank-sum test (p<0.01). (C) Histograms of 124-1m group and control (mt) group. Difference of the two distributions is not significant (p>0.05). (D) Primary neurons transfected with 2’-O-Me oligos with either antisense sequences of miR-124 (O-Me-124-as) or control scrambled miR-124 sequence (O-Me-124-sc) (E) Histograms of 2-O-Me-124-as and group and control group. Difference of the two distributions is significant (p<0.01).
Figure 8. Two forms of miR-124 differ their ability to regulate the Itgb1 3’ UTR
(A) Sequences of two predicted miR-124 binding sites in the Itgb1 3’ UTR and complementarity between the predicted sites and miR-124/miR-124-UU21 (see Fig. 3). The additional 5’ U for miR-124-UU21 is indicated in lowercase. (B) The 3’ UTR of Itgb1 was expressed as the 3’ UTR for a luciferase reporter (Itgb1-WT), and the predicted miR-124 target sites in the reporter were mutated individually (Itgb1-s1m and s2m) or together (Itgb1-s12m).The altered sequences in the Itgb1 s1m and s2m mutations are indicated above the sites in A. Normalized luciferase reporter activity shows that the 3’UTR of Itgb1 was inhibited by cotransfected miR-124 RNA duplex, but not a control siRNA duplex (XASH3). Mutation of either predicted binding site partially reduced miR-124 inhibition, while inhibition of both sites reduced inhibition further. (C) While cotransfected miR-124 inhibited the luciferase activity of Itgb1-WT, but miR-124-UU21 did not. Luciferase activities were assayed 22 hr after transfection. Average of three assays is shown, with standard error indicated. ** P<0.01, Student’s t-test.
Figure 9
Microarray analysis was used to identify mRNAs downregulated in P19 cells transfected with miR-124 or miR-124-UU21 synthetic RNA duplexes (see Fig. 2). (A) Venn diagram with number of mRNAs downregulated by miR-124, miR124a-UU21, or both miRNAs, compared to control siRNA transfections. A detection probability with P<0.01 in the control RNA samples and decreased expression with P <0.01 was used as the threshold to identify downregulated mRNAs. Most regulated mRNAs are downregulated by only one of the two miRNAs. (B) Categories of GO cellular components enriched (P<0.01) among genes downregulated by miR-124 or miR-124-UU21 include components of the cytoskeleton.
Similar articles
- βPix-d promotes tubulin acetylation and neurite outgrowth through a PAK/Stathmin1 signaling pathway.
Kwon Y, Jeon YW, Kwon M, Cho Y, Park D, Shin JE. Kwon Y, et al. PLoS One. 2020 Apr 6;15(4):e0230814. doi: 10.1371/journal.pone.0230814. eCollection 2020. PLoS One. 2020. PMID: 32251425 Free PMC article. - Receptor for advanced glycation end products (RAGE) mediates neuronal differentiation and neurite outgrowth.
Wang L, Li S, Jungalwala FB. Wang L, et al. J Neurosci Res. 2008 May 1;86(6):1254-66. doi: 10.1002/jnr.21578. J Neurosci Res. 2008. PMID: 18058943 - Phosphatidylinositol 3-kinase, Cdc42, and Rac1 act downstream of Ras in integrin-dependent neurite outgrowth in N1E-115 neuroblastoma cells.
Sarner S, Kozma R, Ahmed S, Lim L. Sarner S, et al. Mol Cell Biol. 2000 Jan;20(1):158-72. doi: 10.1128/MCB.20.1.158-172.2000. Mol Cell Biol. 2000. PMID: 10594018 Free PMC article. - miR-744 and miR-224 Downregulate Npas4 and Affect Lineage Differentiation Potential and Neurite Development During Neural Differentiation of Mouse Embryonic Stem Cells.
Choy FC, Klarić TS, Koblar SA, Lewis MD. Choy FC, et al. Mol Neurobiol. 2017 Jul;54(5):3528-3541. doi: 10.1007/s12035-016-9912-4. Epub 2016 May 17. Mol Neurobiol. 2017. PMID: 27189618 - Regulation of neurite outgrowth by G(i/o) signaling pathways.
Bromberg KD, Iyengar R, He JC. Bromberg KD, et al. Front Biosci. 2008 May 1;13:4544-57. doi: 10.2741/3022. Front Biosci. 2008. PMID: 18508528 Free PMC article. Review.
Cited by
- microRNA-124 regulates Notch and NeuroD1 to mediate transition states of neuronal development.
Konrad KD, Song JL. Konrad KD, et al. Dev Neurobiol. 2023 Jan;83(1-2):3-27. doi: 10.1002/dneu.22902. Epub 2022 Nov 23. Dev Neurobiol. 2023. PMID: 36336988 Free PMC article. - RNA processing in neurological tissue: development, aging and disease.
Szeto RA, Tran T, Truong J, Negraes PD, Trujillo CA. Szeto RA, et al. Semin Cell Dev Biol. 2021 Jun;114:57-67. doi: 10.1016/j.semcdb.2020.09.004. Epub 2020 Oct 16. Semin Cell Dev Biol. 2021. PMID: 33077405 Free PMC article. Review. - Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants.
Sun K, Westholm JO, Tsurudome K, Hagen JW, Lu Y, Kohwi M, Betel D, Gao FB, Haghighi AP, Doe CQ, Lai EC. Sun K, et al. PLoS Genet. 2012 Feb;8(2):e1002515. doi: 10.1371/journal.pgen.1002515. Epub 2012 Feb 9. PLoS Genet. 2012. PMID: 22347817 Free PMC article. - Transcriptional dynamics of microRNAs and their targets during Drosophila neurogenesis.
Menzel P, McCorkindale AL, Stefanov SR, Zinzen RP, Meyer IM. Menzel P, et al. RNA Biol. 2019 Jan;16(1):69-81. doi: 10.1080/15476286.2018.1558907. Epub 2019 Jan 20. RNA Biol. 2019. PMID: 30582411 Free PMC article. - MicroRNAs as the cause of schizophrenia in 22q11.2 deletion carriers, and possible implications for idiopathic disease: a mini-review.
Forstner AJ, Degenhardt F, Schratt G, Nöthen MM. Forstner AJ, et al. Front Mol Neurosci. 2013 Dec 5;6:47. doi: 10.3389/fnmol.2013.00047. Front Mol Neurosci. 2013. PMID: 24367288 Free PMC article. Review.
References
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. - PubMed
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. - PubMed
- Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. - PubMed
- Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell. 2008;29:1–7. - PubMed
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous