Molecular basis for the thiol sensitivity of insulin-degrading enzyme - PubMed (original) (raw)
Molecular basis for the thiol sensitivity of insulin-degrading enzyme
Marie Neant-Fery et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Insulin-degrading enzyme (IDE) is a ubiquitous zinc-metalloprotease that hydrolyzes several pathophysiologically relevant peptides, including insulin and the amyloid beta-protein (Abeta). IDE is inhibited irreversibly by compounds that covalently modify cysteine residues, a mechanism that could be operative in the etiology of type 2 diabetes mellitus (DM2) or Alzheimer's disease (AD). However, despite prior investigation, the molecular basis underlying the sensitivity of IDE to thiol-alkylating agents has not been elucidated. To address this topic, we conducted a comprehensive mutational analysis of the 13 cysteine residues within IDE. Our analysis implicates C178, C812, and C819 as the principal residues conferring thiol sensitivity. The involvement of C812 and C819, residues quite distant from the catalytic zinc atom, provides functional evidence that the active site of IDE comprises two separate domains that are operational only in close apposition. Structural analysis and other evidence predict that alkylation of C812 and C819 disrupts substrate binding, whereas alkylation of C178 interferes with the apposition of active-site domains and subtly repositions zinc-binding residues. Unexpectedly, alkylation of C590 was found to activate hydrolysis of Abeta significantly, while having no effect on insulin, demonstrating that chemical modulation of IDE can be both bidirectional and highly substrate selective. Our findings resolve a long-standing riddle about the basic enzymology of IDE with important implications for the etiology of DM2 and AD. Moreover, this work uncovers key details about the mechanistic basis of the unusual substrate selectivity of IDE that may aid the development of pharmacological agents or IDE mutants with therapeutic value.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Cysteine residues present in human IDE. (A) Positions of cysteine residues (red) accessible to the surface of human IDE as viewed from different perspectives. The N- and C-terminal halves of human IDE are shown in green and blue, respectively, and the zinc atom is depicted as a magenta sphere. The modeled portion of the protein, which includes C974, is depicted in light blue. Note the proximity of C812 and C819 (red) to the active-site zinc (magenta) when in the closed conformation (Lower Right). (B) Cysteine residues (yellow) near the active site of IDE. Note the placement of C178 near the junction between the N- and C-terminal halves. The zinc atom is depicted as a gray sphere. Dashed lines show the distance between neighboring residues that could be impacted by alkylation. The distances between the sulfur atom in C178 and the nearest atoms in L116 and T825 are 3.02 Å and 5.69 Å, respectively. The figure was constructed from Protein Data Bank ID code 2G54 (20) by using PyMOL (32).
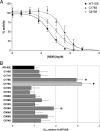
Fig. 2.
Effect of individual C-to-S mutations on NEM-mediated inhibition of IDE. (A) Dose–response curves showing the activity of IDE-C178S, IDE-C819S, and WT-IDE in the presence of varying concentrations of NEM. (B) Graph depicting the IC50 values of individual C-to-S mutants relative to WT-IDE. Data are mean ± SEM for 3–14 replications per condition. *, P < 0.05; ♦, P < 0.001.
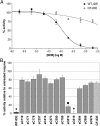
Fig. 3.
Effects of NEM on the activity of CF-IDE and single-C IDE mutants. (A) Dose–response curves showing the activity of WT-IDE and CF-IDE in the presence of varying concentrations of NEM. Note that, despite strong resistance to NEM, CF-IDE is partially inhibited at high concentrations. (B) Graph depicting the activity of single-C mutants in the presence of 2 mM NEM relative to WT-IDE and CF-IDE. Data are mean ± SEM for three to five replications per condition. ●, P < 0.01; ♦, P < 0.001.
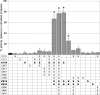
Fig. 4.
Effects of NEM on the activity of IDE mutants containing various C-to-S mutations. Graph shows activity for IDE containing C-to-S mutations at the sites indicated in the table below. Note that resistance to NEM is observed if and only if mutations are present in C178, C812, and C819 simultaneously. Data are mean ± SEM for three to six replications per condition. *, P < 0.05; ♦, P < 0.001.
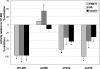
Fig. 5.
Substrate-specific effects induced by alkylation of individual cysteines in IDE. Graph depicts the percentage of activation or inhibition observed by using different single-C mutants and different substrates after treatment with 2 mM NEM. Data are presented as the percentage of the activity of CF-IDE under the same condition (see
Fig. S3
for the raw dataset). Data are mean ± SEM for three to five replications per condition. Statistical significance reflects comparison with the activity of CF-IDE using the same substrate. *, P < 0.05; ●, P < 0.01; ♦, P < 0.001.
Similar articles
- Molecular bases for the recognition of short peptide substrates and cysteine-directed modifications of human insulin-degrading enzyme.
Malito E, Ralat LA, Manolopoulou M, Tsay JL, Wadlington NL, Tang WJ. Malito E, et al. Biochemistry. 2008 Dec 2;47(48):12822-34. doi: 10.1021/bi801192h. Biochemistry. 2008. PMID: 18986166 Free PMC article. - Functional analysis of conserved residues in the active site of insulin-degrading enzyme.
Perlman RK, Gehm BD, Kuo WL, Rosner MR. Perlman RK, et al. J Biol Chem. 1993 Oct 15;268(29):21538-44. J Biol Chem. 1993. PMID: 8104941 - Degradation of Alzheimer's Amyloid-β by a Catalytically Inactive Insulin-Degrading Enzyme.
Sahoo BR, Panda PK, Liang W, Tang WJ, Ahuja R, Ramamoorthy A. Sahoo BR, et al. J Mol Biol. 2021 Jun 25;433(13):166993. doi: 10.1016/j.jmb.2021.166993. Epub 2021 Apr 16. J Mol Biol. 2021. PMID: 33865867 Free PMC article. - Insulin-degrading enzyme: structure-function relationship and its possible roles in health and disease.
Fernández-Gamba A, Leal MC, Morelli L, Castaño EM. Fernández-Gamba A, et al. Curr Pharm Des. 2009;15(31):3644-55. doi: 10.2174/138161209789271799. Curr Pharm Des. 2009. PMID: 19925417 Review. - Impact of Insulin Degrading Enzyme and Neprilysin in Alzheimer's Disease Biology: Characterization of Putative Cognates for Therapeutic Applications.
Jha NK, Jha SK, Kumar D, Kejriwal N, Sharma R, Ambasta RK, Kumar P. Jha NK, et al. J Alzheimers Dis. 2015;48(4):891-917. doi: 10.3233/JAD-150379. J Alzheimers Dis. 2015. PMID: 26444774 Review.
Cited by
- Nontoxic antimicrobials that evade drug resistance.
Davis SA, Vincent BM, Endo MM, Whitesell L, Marchillo K, Andes DR, Lindquist S, Burke MD. Davis SA, et al. Nat Chem Biol. 2015 Jul;11(7):481-7. doi: 10.1038/nchembio.1821. Epub 2015 Jun 1. Nat Chem Biol. 2015. PMID: 26030729 Free PMC article. - Designed inhibitors of insulin-degrading enzyme regulate the catabolism and activity of insulin.
Leissring MA, Malito E, Hedouin S, Reinstatler L, Sahara T, Abdul-Hay SO, Choudhry S, Maharvi GM, Fauq AH, Huzarska M, May PS, Choi S, Logan TP, Turk BE, Cantley LC, Manolopoulou M, Tang WJ, Stein RL, Cuny GD, Selkoe DJ. Leissring MA, et al. PLoS One. 2010 May 7;5(5):e10504. doi: 10.1371/journal.pone.0010504. PLoS One. 2010. PMID: 20498699 Free PMC article. - Development and Characterization of Quantitative, High-Throughput-Compatible Assays for Proteolytic Degradation of Glucagon.
Suire CN, Lane S, Leissring MA. Suire CN, et al. SLAS Discov. 2018 Dec;23(10):1060-1069. doi: 10.1177/2472555218786509. Epub 2018 Jul 11. SLAS Discov. 2018. PMID: 29995452 Free PMC article. - Peptidic inhibitors of insulin-degrading enzyme with potential for dermatological applications discovered via phage display.
Suire CN, Nainar S, Fazio M, Kreutzer AG, Paymozd-Yazdi T, Topper CL, Thompson CR, Leissring MA. Suire CN, et al. PLoS One. 2018 Feb 15;13(2):e0193101. doi: 10.1371/journal.pone.0193101. eCollection 2018. PLoS One. 2018. PMID: 29447281 Free PMC article. - Cysteine 904 is required for maximal insulin degrading enzyme activity and polyanion activation.
Song ES, Melikishvili M, Fried MG, Juliano MA, Juliano L, Rodgers DW, Hersh LB. Song ES, et al. PLoS One. 2012;7(10):e46790. doi: 10.1371/journal.pone.0046790. Epub 2012 Oct 15. PLoS One. 2012. PMID: 23077523 Free PMC article.
References
- Tanzi RE, Moir RD, Wagner SL. Clearance of Alzheimer's Aβ peptide: The many roads to perdition. Neuron. 2004;43:605–608. - PubMed
- Leissring MA, Saido TC. In: Alzheimer's Disease: Advances in Genetics, Molecular and Cellular Biology. Sisodia S, Tanzi R, editors. New York: Springer; 2007. pp. 157–178.
- Bertram L, et al. Evidence for genetic linkage of Alzheimer's disease to chromosome 10q. Science. 2000;290:2302–2303. - PubMed
- Myers A, et al. Susceptibility locus for Alzheimer's disease on chromosome 10. Science. 2000;290:2304–2305. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources